Fig. 8.
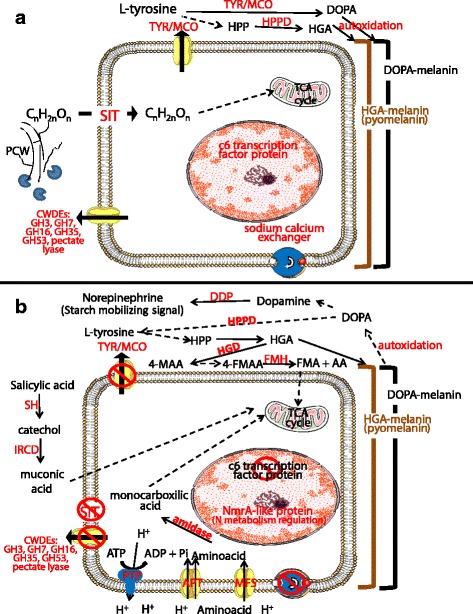
Hypothetical model of in vitro L. theobromae heat stress (HS) response, in the presence of grapevine wood (GW). The model of whole cell response was constructed using genes from clusters 1, 3 and 7 (in Fig. 2) showing both co-regulation and GO term enrichment. Additional file 13 gives the gene name with its expression pattern. The illustration shows the main functions and processes being carried out by the fungus when growing on GW (a), and the changes that are proposed to be occurring after HS (b). The putative cellular location (intra or extracellular) is indicated in the scheme. A continuous arrow line indicates an enzymatic reaction that is supported on the differentially expression of its encoding gene (shown in red). A dashed arrow line indicates a suggested enzymatic reaction. The substrate/product of reaction is indicated on boxes. TYR: tyrosinase, MCO: multicopper oxidase, DOPA: L-3,4-dihydroxyphenylalanine, HPP: 4-hydroxyphenylpyruvate, HPPD: 4-hydroxyphenylpyruvate dioxygenase, HGD: homogentisate dioxygenase, HGA: homogentisic acid, 4-MAA: 4-maleylacetoacetate, 4-FMAA: 4-fumarylacetoacetate, FM: fumarate, AA: acetoacetate, FMH:fumarylacetoacetate hydrolase, SH: salicylate hydroxylase, IRCD: intradiol ring cleavage dioxygenase, DDP: domon domain containing protein, PCWDEs: Plant cell wall degrading enzymes; PCW: Plant cell wall
