Figure 1.
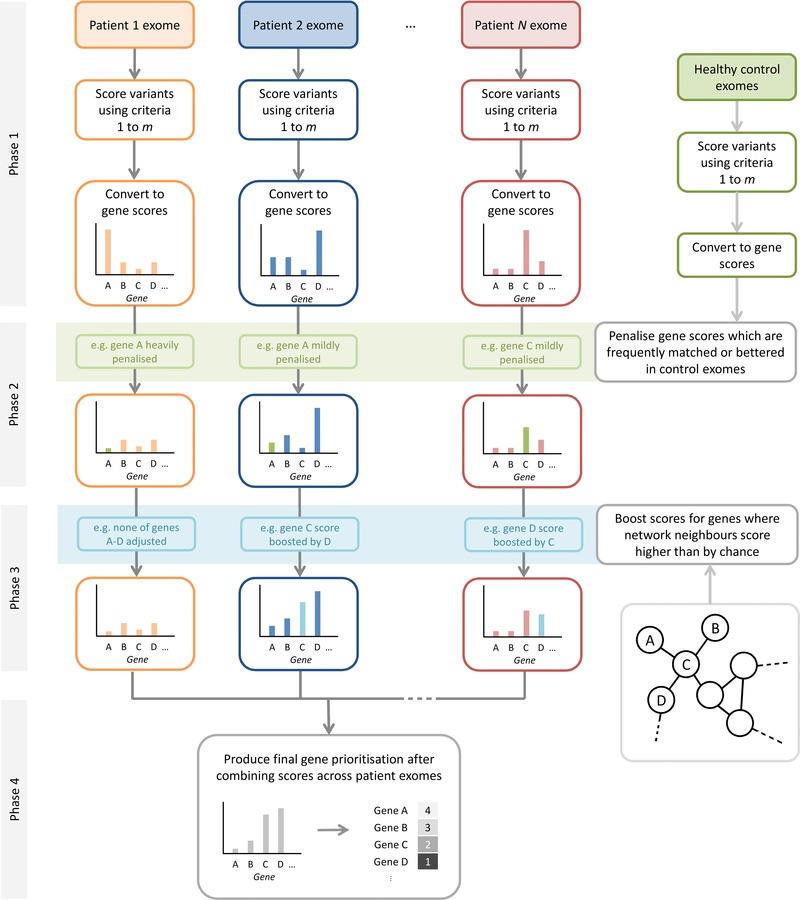
The HetRank analysis framework. Phase 1: Variants are ranked in each affected individual's exome sequence according to a set of user‐specified criteria and converted into gene scores. Phase 2: Gene scores are adjusted with respect to the scores achieved in a set of healthy control exomes, allowing more accurate prioritization of long and variant‐tolerating genes. Phase 3: An interaction network is used to share score information between neighboring genes. Neighboring genes that score highly in different affected individuals improve the evidence for one another's involvement in the disease process. Phase 4: Scores are combined across all exomes in the study to give a final prioritized list of genes.
