FIGURE 1.
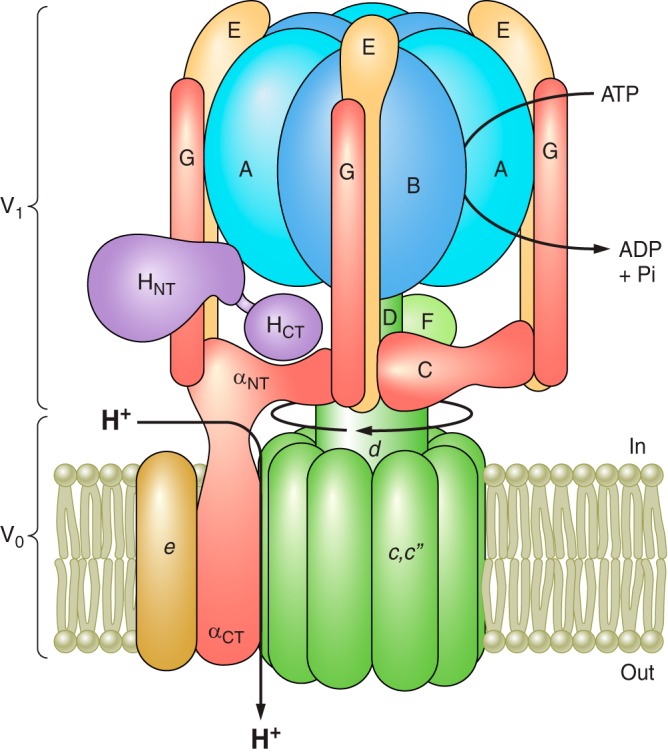
Structure and mechanism of the V-ATPase. The V-ATPase is composed of a peripheral V1 domain that hydrolyzes ATP and an integral V0 domain that translocates protons. ATP hydrolysis occurs at nucleotide binding sites located at the interface of the A and B subunits and drives rotation of a central rotary complex composed of subunits D and F of V1 and subunit d and the ring of proteolipid subunits (c and c”) of V0 relative to the remainder of the complex. Rotation of the proteolipid ring relative to subunit a drives unidirectional proton transport from the cytoplasm to the lumen (see text for details). The A3B3 catalytic head is held fixed relative to subunit a by peripheral stalks composed of three EG heterodimers that connect to subunits C and H and the NH2-terminal cytoplasmic domain of subunit a. Model is adapted from Couoh-Cardel et al. (26). See Reference 232 for a recent model based on cryo-EM of the yeast V-ATPase.
