Figure 1.
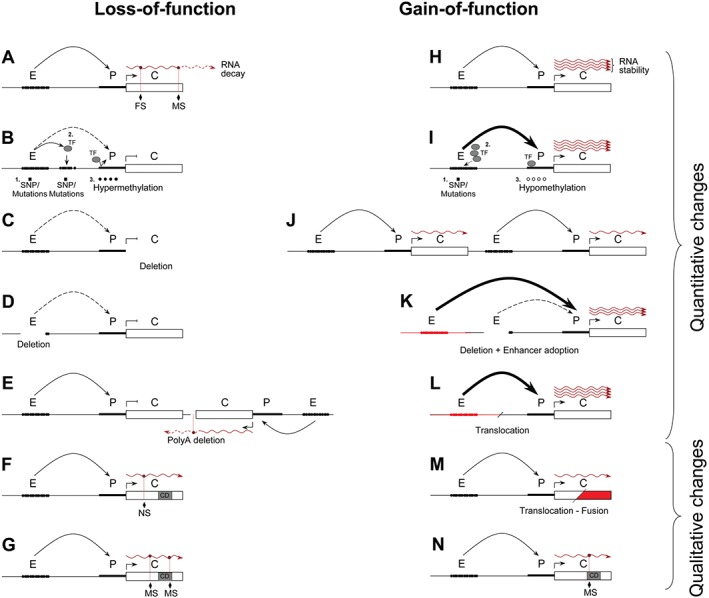
Molecular basis of genetic diseases. Effects of loss‐ and gain‐of‐function mutations affecting gene expression are quantitative and/or qualitative. (A) A missense mutation or a small insertion/deletion mutation (frameshift) in a coding sequence or at a PolyA signal often leads to abortive translation or RNA decay 162. (B) Reduction of chromosomal looping between the enhancer and the promoter might be due to (1) natural variant or mutation at the enhancer 163, (2) the presence of a new SNP forming a new enhancer/promoter region which titrates the remote enhancer activity 43, or (3) promoter or enhancer hypermethylation 164. (C) Deletion of the gene 165. (D) Deletion of the remote enhancer 166. (E) Deletion of the PolyA signal of a downstream and convergent gene, leading to the production of antisense RNA 167. (F) Nonsense mutation adding a new premature stop codon producing a truncated protein 168. Note that truncated proteins may also have a gain‐of‐function activity 169. (G) Missense mutation affecting the non‐enzymatic activity or abolishing the catalytic domain of an enzyme 104. (H) Normal rate of transcription, but increased accumulation of final gene product due to the presence of an RNA 170 or a protein 171 stabilizing molecule. (I) Increased enhancer activity due to (1) enhancer mutation 25, (2) overexpression of a transcription factor 172, or (3) promoter hypomethylation 173. (J) An increase in gene copy number, including regulatory regions 174. (K) Large genomic deletion bringing a strong (but irrelevant) enhancer closer 175. (L) Translocation with a heterologous chromosome (red) creating a fusion locus with a new strong enhancer regulating an illegitimate gene 176. (M) Translocation with a heterologous chromosome (red) producing a fusion gene, with increased biological activity 96. (N) Missense mutation improving enzymatic activity 81. E, enhancer; P, promoter; C, coding region; TF, transcription factor; CD, catalytic domain; MS, missense mutation; NS, nonsense mutation; FS, frameshift mutation. Dashed curved arrows represent impaired enhancer–promoter interaction (looping); thin curved arrows, normal looping; and thick curved arrows, strong looping. Wavy red lines indicate mRNA.
