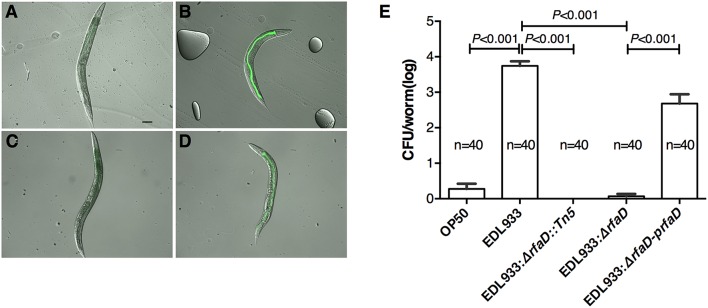
Figure 3.
Mutation in rfaD reduces EDL933 colonization in C. elegans. Images of wild-type N2 nematodes fed with GFP-labeled OP50 (A), EDL933 (B), EDL933: ΔrfaD (C) or EDL933:ΔrfaD-prfaD (D) for 1 days at 20°C and then chased with non GFP-labeled OP50 for 3 days at 20°C, respectively. Animals previously exposed on GFP-labeled EDL933 and EDL933:ΔrfaD-prfaD plates showed significant GFP signals in their intestines. Animals fed with wild-type EDL933 and EDL933:ΔrfaD-prfaD exhibited unhealthy apperances with smaller and paler body compared to animals fed with OP50 and EDL933:ΔrfaD. Representative images are shown, and the scale bar represents 100 μm. (E) The number of bacteria colonized in C. elegans was determined by the colony forming units (CFU) assay. Values represent the means of three independent assays, and error bars indicate the standard deviations. P-values denote the results of statistical analysis. The total numbers of animals tested in each group are indicated by n.