Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
Upon in vitro differentiation, iPSCs obtained from patients with SCID and OS show a similar block in T-cell development.
Presence of unresolved single-strand DNA breaks in developing T cells from OS patient-derived iPSCs affects their differentiation.
Abstract
Primary immunodeficiency diseases comprise a group of heterogeneous genetic defects that affect immune system development and/or function. Here we use in vitro differentiation of human induced pluripotent stem cells (iPSCs) generated from patients with different recombination-activating gene 1 (RAG1) mutations to assess T-cell development and T-cell receptor (TCR) V(D)J recombination. RAG1-mutants from severe combined immunodeficient (SCID) patient cells showed a failure to sustain progression beyond the CD3−-CD4−CD8−CD7+CD5+CD38−CD31−/loCD45RA+ stage of T-cell development to reach the CD3−/+CD4+CD8+CD7+CD5+CD38+CD31+CD45RA− stage. Despite residual mutant RAG1 recombination activity from an Omenn syndrome (OS) patient, similar impaired T-cell differentiation was observed, due to increased single-strand DNA breaks that likely occur due to heterodimers consisting of both an N-terminal truncated and a catalytically dead RAG1. Furthermore, deep-sequencing analysis of TCR-β (TRB) and TCR-α (TRA) rearrangements of CD3−CD4+CD8− immature single-positive and CD3+CD4+CD8+ double-positive cells showed severe restriction of repertoire diversity with preferential usage of few Variable, Diversity, and Joining genes, and skewed length distribution of the TRB and TRA complementary determining region 3 sequences from SCID and OS iPSC-derived cells, whereas control iPSCs yielded T-cell progenitors with a broadly diversified repertoire. Finally, no TRA/δ excision circles (TRECs), a marker of TRA/δ locus rearrangements, were detected in SCID and OS-derived T-lineage cells, consistent with a pre-TCR block in T-cell development. This study compares human T-cell development of SCID vs OS patients, and elucidates important differences that help to explain the wide range of immunologic phenotypes that result from different mutations within the same gene of various patients.
Introduction
Primary immunodeficiency diseases (PIDs) include over 200 distinct disorders that variably influence immune system development and function, thereby causing increased susceptibility to infections, autoimmunity, and malignancies.1 In particular, severe combined immunodeficiency (SCID) disorders are characterized by a block in T-cell development, variably associated with impaired B and/or natural killer (NK) cell differentiation. Mechanisms that account for SCID include impaired cell survival (adenosine deaminase deficiency, reticular dysgenesis), defective response to cytokines (IL7R, IL2RG, and JAK3 gene defects), impaired assembly of the antigen receptor (recombination-activating gene 1/2 (RAG1/2), DCLRE1C, PRKDC, and LIG4 defects), and defective signaling through the pre–T-cell receptor (TCR) and/or the TCR (defects of CD3δ, CD3ε, CD3ξ, ZAP70, and CD45). Severe mutations of these genes block T-cell development prior to differentiation of single-positive thymocytes.
The RAG1 and RAG2 proteins initiate the process of Variable (V), Diversity (D), and Joining (J) gene recombination by recognizing recombination signal sequences (RSS) that flank the V-D-J gene elements within the TCR and immunoglobulin (Ig) gene loci.2 RAGs first nick a single DNA strand, which allows them to introduce DNA double-strand breaks (DNA-DSBs) that are initially covalently sealed by a hairpin (coding ends [CEs]).3 Subsequently, the DNA-PK catalytic subunit (DNA-PKcs) protein activates DNA cross-link repair 1C (DCLRE1C; also known as Artemis), allowing opening of the hairpin. The DNA-DSBs are then repaired by proteins of the nonhomologous end-joining pathway, thereby permitting the juxtaposition of nonadjacent V-D-J genes.4 RAG mutations in humans are associated with a variety of clinical and immunologic phenotypes that reflect the biochemical consequences of the mutation and the effect of environmental factors.5 In patients with null RAG mutations, complete failure of V(D)J recombination is associated with complete lack of circulating T and B lymphocytes, hence resulting in the T− B− NK+ form of SCID. We and others have shown that hypomorphic RAG mutations that affect, but do not abolish, V(D)J recombination, are often associated with distinct immunologic and clinical phenotypes with residual presence of T, and in some cases B, lymphocytes.6-9 The presence of autologous, auto-reactive, activated, and oligoclonal T cells that infiltrate and damage peripheral organs is a hallmark of Omenn syndrome (OS). In other cases, hypomorphic RAG mutations may cause delayed disease onset, granuloma formation, autoimmunity, and/or dysgammaglobulinemia.5
Using an in vitro cellular platform in which RAG activity can be measured by analyzing recombination at an inverted green fluorescent protein (GFP) cassette flanked by RSS, we have shown that the phenotypic diversity of human RAG deficiency correlates with the residual function of the mutant RAG protein.10 We found that mutations associated with OS have residual, yet markedly decreased, recombination activity. The observation that OS and T− B− NK+ SCID may occur in affected members of the same family suggests that RAG mutations associated with these phenotypes can only support, at best, limited repertoire diversity. However, no studies have compared T-cell development in patients with RAG mutations associated with OS vs SCID.
Mouse models have been used to elucidate the functions of genes involved in PID, and SCID in particular. A mouse model for SCID was first reported by Bosma et al,11 the result of a naturally occurring mutation in the Prkdc gene.12 Although the scid mouse is initially deficient in functional T and B cells, some young adult mice generate a low number of functional lymphocytes, and a leaky SCID phenotype is observed in most scid mice by 1 year.13 In contrast, the Rag1 or Rag2 null mice result in a nonleaky SCID, with a stringent block at the CD4−CD8− CD44−CD25+ double negative 3 stage of intrathymic T-cell development, resulting in absence of B or T lymphocytes.14,15 Mouse models of OS and of leaky SCID have been generated, such as Rag1 R972Q,16 the Rag1 S723C,7, and Rag2 R229Q17 mice. In addition to the mouse, SCID and SCID variants have also been modeled in the dog and horse.18,19
Although animal models serve as an important tool for elucidating gene functions, and how certain mutations result in PIDs, there is a clear need to study PIDs in a human context. There are differences in T-lymphocyte development between humans and mice,20 and disease mechanisms likely differ as well. However, several obstacles exist that make it difficult to study the developmental pathophysiology of human SCID at the cellular and molecular level, including rarity of the disease, the urgency of treatment, and difficulties in obtaining appropriate tissue samples. Recent work has demonstrated that T cells can be generated from human induced pluripotent stem cells (iPSCs) in vitro.21-23 This in vitro approach can reduce the need for using animal models in place of a more ethical, rapid, and more cost-effective means to conduct research within a human context, validating treatment or the repair of a patient’s defective gene in the context of thymocyte differentiation. A first report that defective T-cell differentiation associated with SCID can be modeled using patient-derived iPSCs has been provided by demonstrating an early arrest of T-cell development of cells carrying an IL2RG mutation, responsible for X-linked SCID, and rescue of T-cell differentiation by means of transcription activator-like effector nucleases-mediated gene editing.24 Although recent work in the field adds to the understanding of PIDs in a human context, they were not aimed at elucidating the causality of the broad spectrum of clinical and immunologic phenotypes resulting from different mutations within the same gene. We reprogrammed dermal fibroblasts to generate iPSCs from patients with RAG1 mutations that resulted in different clinical and immunologic phenotypes (SCID and OS). When control and patient-derived iPSCs were differentiated toward the T-cell lineage in vitro, a primary block in T-cell development at an early stage (CD7+CD5− or CD7+CD5+ stage) was observed both in OS and in SCID cells, respectively. In both instances, developmental progression was briefly observed up to the CD4+CD8− immature single-positive (ISP) and CD4+CD8+ double-positive (DP) stage, but was not sustained, which correlated with the lack of intracellular TCR-β (TRB) for SCID and OS cells, and increased detection of single-strand DNA breaks unique to OS cells. Additionally, assessment of TRB and TCR-α (TRA) recombination within the SCID and OS-derived ISP and DP populations showed limited repertoire diversity, with skewed usage of individual V-D-J genes, abnormal distribution, and length of the TRB and TRA complementary determining region 3 (CDR3), and an absence of TRA/δ excision circles (TRECs). Overall, these results confirm that RAG mutations associated with OS and SCID are similarly severe, with key differences unique to OS cells that help to explain the broad phenotypic diversity of RAG1-associated PIDs, and demonstrate the applicability of using iPSCs to model PID and to study mechanisms of altered T-cell development in humans.
Methods
Patients
Three patients with bi-allelic RAG1 mutations were included in the study. Patient 1 (P1) was born to first cousin parents, and suffered from recurrent lower respiratory tract infections since the first month of life. Laboratory tests performed at 5 months of age demonstrated severe lymphopenia (140 cells/μL). Lymphocyte subsets were as follows: CD3: 1.4%; CD4: 0.3%; CD8: 0.1%; CD19: 1.1%; and CD16: 74.2%. Serum IgA (1 mg/dL) and IgM (2 mg/dL) were markedly decreased, whereas IgG serum levels (385 mg/dL) likely reflected maternally transferred IgG. Molecular analysis revealed a homozygous RAG1 mutation (c.1428delC), predicted to result in frameshift and premature termination (p.N476Kfs*16).
P2 was a Kuwaiti infant born to consanguineous (first cousin) parents. She presented with diarrhea, growth arrest, recurrent respiratory tract infections, and oral candidiasis since the first months of life. At 11 months of age, the absolute lymphocyte count was 1848 cells/μL, with abnormal distribution of lymphocyte subsets (CD3: 15%; CD4: 14%; CD8: 0%; CD19: 0.5%; and CD16: 79%). In vitro proliferation to phytohemagglutinin was markedly decreased (2700 cpm vs 166 000 in a healthy control). Molecular analysis revealed a homozygous RAG1 mutation (c.1180C>T), resulting in the R394W amino acid substitution.
P3, of Slavic origin, was born to nonconsanguineous parents. An older sibling had been diagnosed with OS and had died of complications after hematopoietic stem cell transplantation. The patient developed generalized skin erythema, lymphadenopathy, and hepatomegaly at 2 months of age. Laboratory tests demonstrated leukocytosis (white blood cells, 46 100/μL), lymphocytosis (18 300 cells/μL), and eosinophilia (7039 cells/mL). Lymphocyte subsets were as follows: CD3: 91.9% and CD4: 62.4% (98.8% of which were CD45R0+); CD8: 27.6%; CD19: 0.2%; and CD16: 3.9%. Hypogammaglobulinemia (IgG: 245 mg/dL) with markedly elevated IgE (>1000 kU/L) were demonstrated. In vitro proliferation to phytohemagglutinin was nearly abolished (2000 cpm vs 172 000 in a healthy control). Maternal T-cell engraftment was ruled out by microsatellite analysis. Molecular analysis demonstrated compound heterozygous RAG1 mutations (c.256_257del; c.2164G>A), predicted to cause the K86Vfs*33 and E722K protein changes, respectively.
On the basis of the Primary Immune Deficiency Treatment Consortium criteria,25 P1 and P2 met the definition of SCID, whereas P3 was diagnosed with OS.
Online materials
See supplemental Methods, available on the Blood Web site. Online material includes methods for: (1) analysis of recombination activity of the mutant RAG1 proteins; (2) generation and characterization of RAG1-deficient iPSCs; (3) iPSC maintenance and differentiation; 4) CD34+ isolation; (5) OP9-DL-4 coculture and differentiation; (6) flow cytometry; (7) next-generation sequencing of TRB and TRA repertoires; (8) Quantification of TRECs; (9) single cell gel electrophoresis (Comet assay); (10) Southern blot analysis; (11) protein expression and purification for in vitro analysis; (12) in vitro DNA nicking assays; (13) in vitro binding assays; and (14) statistical analysis.
Results
RAG1 mutations of 3 patients have low recombination activity and result in cleavage defects
In order to investigate the effects of RAG1 deficiency on human T-cell development, we generated iPSCs from dermal fibroblasts obtained from 2 patients with SCID and 1 patient with OS. P1 and P2 with SCID were both homozygous for distinct RAG1 mutations, whereas compound heterozygosity was demonstrated in P3 with OS (Figure 1). The SCID P1 single nucleotide deletion at residue 1428 is predicted to result in frameshift and early truncation of the RAG1 protein with only the nonamer-binding region (NBR) domain of the catalytic core intact. The SCID P2 and OS P3 substitution mutations within the NBR or heptamer-binding region respectively, are predicted to severely limit recognition and/or binding of the RSS. The OS P3 deletion (c.256_257delAA) was previously reported to produce an N-terminal truncated RAG1 protein with an intact catalytic domain, due to use of an alternative start codon; this mutant is severely blocked from entering the nucleus.26 To characterize the levels of recombination activity of each of these RAG1 mutants, Abelson-transformed Rag1−/− pro–B-cells containing an inverted GFP cassette flanked by RSS (pMX-INV cassette) were transduced with retroviral vectors encoding for the various RAG1 mutant proteins or wild-type (WT) RAG1, and GFP expression was used as a read-out of the recombination activity.10 Total cell lysates were obtained for immunoblot analysis in order to assess RAG1 expression levels (Figure 2A). The antibody used (D36B3; Cell Signaling) is N-terminus specific and therefore does not recognize the product of the c.256_257delAA mutation.
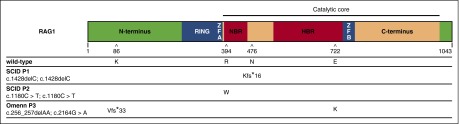
Figure 1.
Scheme of RAG1 protein and mutations. WT human RAG1 consists of 1043 amino acids and includes an N-terminus domain, really interesting new gene (RING) finger sequence, zinc finger sequences zinc finger A (ZFA) and zinc finger B (ZFB), the catalytic core, which contains the nonamer- and heptamer-binding regions, and a C-terminus domain. The amino acid positions affected by mutations identified in patients with SCID (P1 and P2) and with OS (P3), and the respective consequences on amino acid sequence are shown. P1 and P2 were homozygous for a frameshift (N476Kfs*16) and a missense (R394W) mutation, respectively. P3 was compound heterozygous for a missense (E722K) and a frameshift (K86Vfs*33). For the latter, an alternative start codon can be used resulting in an N-terminus truncated protein with normal sequence from Met183 onward with cytoplasmic localization. HBR, heptamer-binding region; NBR, nonamer-binding region.
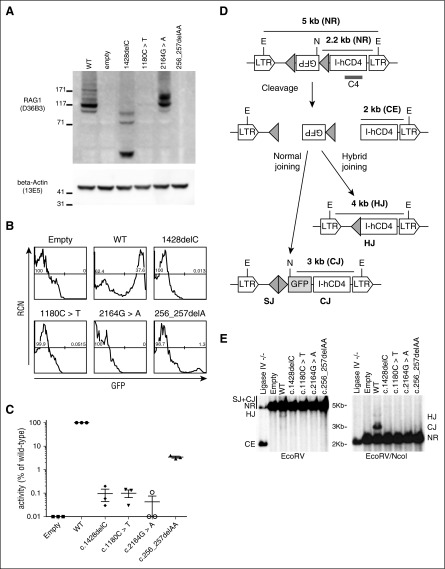
Figure 2.
Recombination activity of mutant RAG1. (A) Immunoblot analysis of Rag1−/− pro-B Abelson cells expressing WT human RAG1, empty vector, or hypomorphic RAG1-mutant alleles from P1 (1428delC), P2 (1180C>T), and P3 (256_257delAA and 2164G>A). N-terminus–specific anti-RAG1 antibody (D36B3) cannot detect the N-terminus–truncated 256_257delAA. β-Actin serves as a loading control, and molecular weight markers (in kDa) are marked. (B) Flow cytometric analysis of Rag1−/− pro-B Abelson cells containing a pMX-INV cassette with an inverted GFP sequence flanked by RSS (shown as gray triangles in [D]), and transduced with retroviral vectors expressing WT, empty, or hypomorphic RAG1 mutants. GFP+ events indicate rearrangement. (C) Recombination activity in transduced Rag1−/− pro-B cells, expressed as a percentage of activity detected in cells expressing WT RAG1. For each construct, 3 independent experiments were performed, and mean value ± standard deviation is shown. (D) Schematic representation showing nonrearranged (NR) products flanked by Escherichia coli RV (EcoRV) (E) sites (5 kb), or EcoRV (E) and Nocardia corallina I (NcoI) (N) sites (2.2 kb). Recombination results in normal joining, producing a 3 kb fragment (coding joining [CJ]) flanked by N and E sites, or hybrid joining (HJ) flanked by V sites and producing a 4 kb fragment. Successful cleavage but unresolved CEs produce a 2 kb fragment. C4 represents the location of the probe to visualize the various fragments for Southern blotting. (E) Southern blot showing products of rearrangement of the pMX-INV vector in pro-B Abelson cells expressing ligase IV null, empty vector, WT RAG1, or one of the four hypomorphic RAG1 mutants. Hybridization with the C4 probe with EcoRV digest reveals NR, HJ, or CE events, and EcoRV+NcoI digest reveals HJ, CJ, or NR events. LTR, long terminal repeat; RCN, relative cell number; SJ, signal joining.
As shown in Figure 2B-C, the RAG1 mutations associated with SCID supported extremely low recombination activity when compared with WT RAG1 (P1, c.1428delC: 0.10 ± 0.05%; P2, c.1180C>T: 0.10 ± 0.04%). One of the 2 mutant alleles associated with OS in P3 (c.2164G>A) had virtually undetectable recombination activity (0.04% ± 0.03), whereas the other allele (256_257delAA) had residual, though markedly decreased, function (3.48 ± 0.35%). These data are consistent with the previous demonstration that the 256_257delAA frameshift mutation results in a prematurely truncated, catalytically inactive RAG1 mutant protein, but use of an alternative downstream translation start site permits expression of an N-terminally truncated RAG1 protein with low recombination activity.26
RAG1 catalyzes recombination of the V-D-J genes through cleaving and rejoining of the TCR locus. To establish whether the RAG1 mutations identified in patients lead to unresolved, unjoined DNA breaks, we analyzed the Abelson pro–B-cells expressing mutant RAG1 (Figure 2D-E). As expected, Southern-blot analysis of the pro-B Abelson cells reconstituted with WT RAG1 revealed a band of the expected size for the rearranged pMX-INV cassette (Figure 2E). Conversely, the RAG1 mutations did not support the generation of detectable levels of pMX-INV rearrangement, with the exception of the c.256_257delAA mutant, for which a very faint band corresponding to rearranged pMX-INV was observed (Figure 2E), reflecting very low levels of recombination observed in OS patients. These results are consistent with the residual recombination activity of this mutant demonstrated by flow cytometry analysis of GFP expression (Figure 2B-C). Additionally, none of the RAG1-reconstituted samples showed accumulation of unrepaired CEs, which are easily detected in a ligase IV−/− cell line, thus confirming that the RAG1 mutants have defects in DNA cleavage.
Generation of CD34+ hematopoietic progenitor cells from RAG1-mutant iPSCs
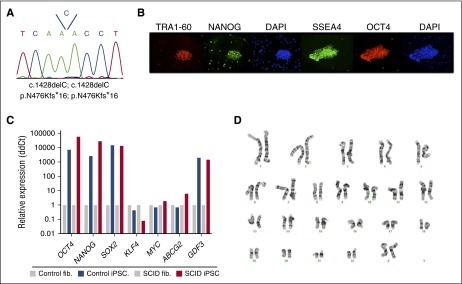
In order to study the developmental block in T-cell development associated with different RAG1 mutants, we took advantage of iPSCs that we had previously generated, starting from dermal fibroblasts of SCID P2 and OS P3,27 as well as of a control iPSC line that had been derived from a healthy control.28 To generate iPSCs from SCID P1, we transduced fibroblasts with a lentiviral vector expressing the 4 Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC). The resulting iPSC colonies were manually picked and expanded. Mutation analysis at the RAG1 locus confirmed their patient-specific origin (Figure 3A). Immunohistochemistry (Figure 3B) and real-time polymerase chain reaction (PCR) (Figure 3C) demonstrated expression of pluripotency and stemness markers, as well as silencing of the KLF4 and c-MYC transgenes. Finally, cytogenetic analysis demonstrated a lack of karyotypic abnormalities (Figure 3D).
Figure 3.
Characterization of an iPSC line reprogrammed from fibroblasts from P1. (A) Sequencing of genomic DNA from the iPSC line, revealing homozygosity for the same single nucleotide deletion at position 1428 identified in parental fibroblasts, and resulting in the p.N476Kfs*16 mutation. (B) Immunofluorescent staining and (C) quantitative PCR analysis of pluripotency markers expressed by the iPSCs. (D) Karyotypic integrity of P1 iPSC line.
To generate CD34+ hematopoietic progenitor cells, control- and RAG1-mutated iPSCs were allowed to differentiate into embryoid bodies,21 and CD34+ progenitor cells were isolated by magnetic-activated cell separation after 8 to 13 days (supplemental Figure 1).
T-cell development analysis from control- and patient-derived iPSC lines
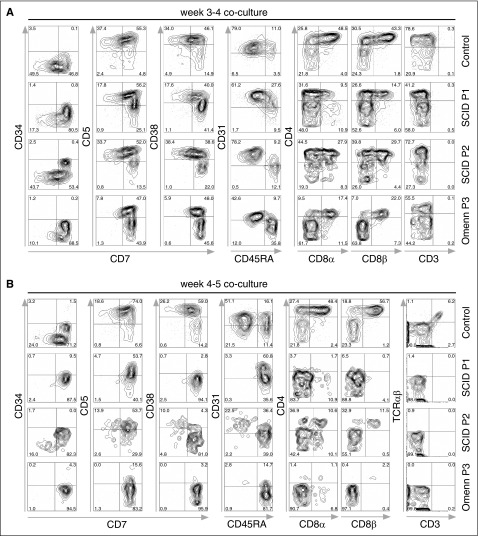
To analyze T-cell developmental progression of control and RAG1-mutated iPSCs, embryoid body-derived CD34+ cells were transferred onto OP9-DL-4 cells in the presence of appropriate cytokines and growth factors to induce T-lineage differentiation. Flow cytometric analyses after 3 to 4 weeks of coculture showed that control, SCID, and OS-derived cells had gained CD7 expression, indicative of T-lineage differentiation (Figure 4A). Additionally, nearly all the cells had downregulated CD34 expression, whereas a majority of the cells also expressed CD5 and CD38. However, a larger proportion of SCID and OS cells were CD7+ CD5− or CD7+ CD38− as compared with control cells.
Figure 4.
In vitro T-lineage differentiation of control and SCID iPSC lines. Flow cytometric analysis of T-lineage developmental progression of control- and patient-derived cells. iPSCS were allowed to differentiate for 8 days into embryoid bodies, and magnetic bead-purified CD34+ cells were cocultured with OP9-DL-4 cells. (A) Cells from P1 and P2 with SCID, and from P3 with OS attained normal expression of early markers of T-lineage differentiation (CD7, CD5, and CD38) upon 3 to 4 weeks of coculture with OP9-DL-4 cells. (B) After 4 to 5 weeks of coculture, cells from a healthy control progress to the CD4+ CD8αβ+ DP stage of differentiation, with the appearance of CD3+ TRA/TRB+ cells. By contrast, SCID- and OS-derived cells were mostly blocked at the CD7+ CD31−/+ CD45RA+ stage of differentiation, with a virtual absence of CD4 and CD8α/β expression, and lack of CD3+ cells. In (A-B), cells were pre-gated for lymphocytes (SSCxFSC), DAPI-, and CD45+. DAPI, 4′,6-diamidino-2-phenylindole; FSC, forward scatter; SSC, side scatter.
Further analysis showed that a larger proportion of RAG1-mutated cells remained CD45RA+ as compared with what was observed in control cells. Although both control and RAG1-mutated cells reached a CD4+ stage of differentiation, a larger proportion of control cells expressed CD4, with nearly half also expressing CD8α and CD8β. In contrast, fewer CD4+ cells were obtained in culture of SCID P1 and OS cells, with only ∼10% to 30% also expressing CD8α and CD8β (Figure 4A). At this time point, neither control nor RAG1-mutated cells showed CD3 expression at the cell surface.
Within 4 to 5 weeks of coculture, as expected, both control and RAG1-mutated cells expressed CD7 and CD5 (Figure 4B). Control cells showed a sustained developmental progression, as illustrated by expression of CD38 and by reaching the CD31+ CD45RA− stage of T-cell differentiation, which also includes expression of CD4 and CD8α/β. By contrast, SCID- or OS-derived cells did not co-express CD38 and CD7, and were mostly blocked at the CD31−/+ CD45RA+ stage of differentiation, with a virtual absence of CD4 and CD8α/β expression. Cell surface expression of CD3 was observed in control cells, but not in SCID- or OS-derived cells. Additionally, intracellular staining for TRB expression showed that 31% of control cells had a productively rearranged TCR, whereas virtually no TCR expression could be detected in SCID or OS cells (supplemental Figure 2). These results suggest that although an initial progression to a CD45RA− CD31+ CD4+ CD8α/β+ stage of differentiation can be observed in RAG1-mutated CD34+ cells cultured on OP9-DL-4 cells, this is not sustained at later time points, with the cells showing the more expected block in T-cell differentiation. Similar results were obtained in at least 3 independent experiments for each iPSC line (supplemental Figure 3A-K). To assess the extent of apoptosis/cell death, cells were stained with Annexin V and SYTOX Green (supplemental Figure 3L), and showed a similar amount of Annexin V-positive staining for each cell line.
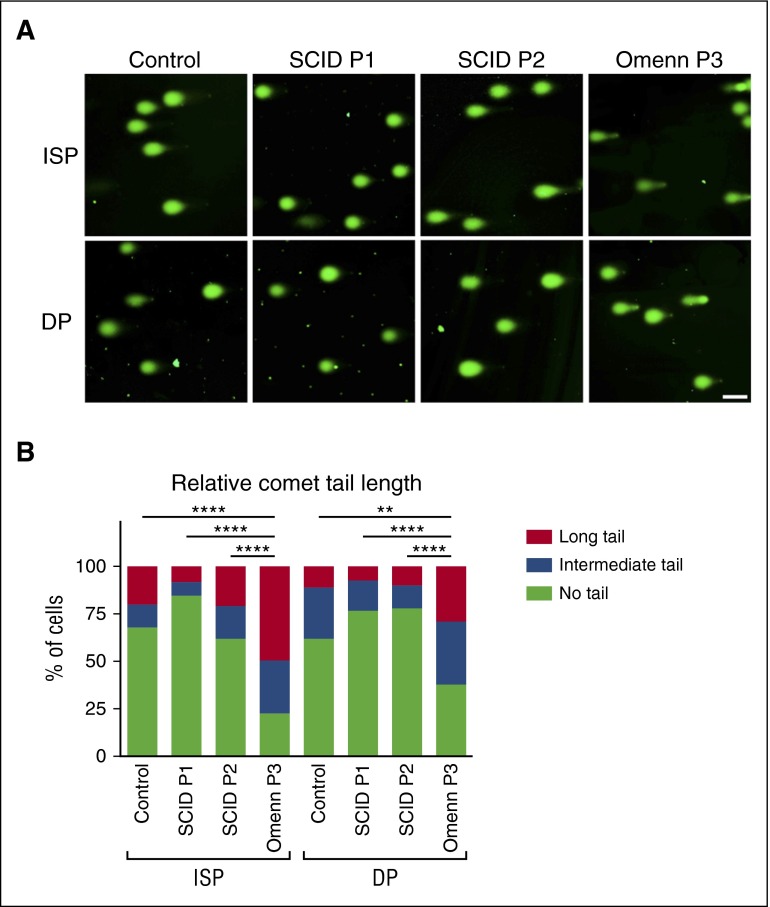
OS cells have a higher propensity for DNA breaks
In order to investigate the potential cause for the lack of sustained developmental progression observed in OS cells, an alkaline single cell gel electrophoresis (Comet assay) was performed,6,29 that would allow to detect DNA nicks or breaks, which relax and open regions of the affected supercoiled DNA resulting in trailing tails (resembling comets) of DNA. Comets were consistently observed in OS cells sorted from coculture day 25 (immediately prior to the block observed for SCID or OS-derived cells), whereas these were absent in equivalent control- or SCID-derived cells (Figure 5A-B). To determine whether SCID- or OS-associated mutations within the catalytic core of RAG1 allow the production of mutant protein capable of DNA nicking, we performed an in vitro oligonucleotide cleavage assay (supplemental Figure 4A). No nicking activity was observed for either R394W (SCID) or E722K (OS) mutants (supplemental Figure 4B). The p.N476Kfs*16 mutant of SCID P1 lacks the catalytic core, and therefore it was not possible (nor necessary) to assay its activity. By contrast, the c.256_257delAA mutant allele of OS P3 is predicted to produce an N-terminus truncated protein (due to usage of a downstream inframe translation initiation site), with an intact catalytic domain.26 Consistent with this, this mutant retained nicking activity, as revealed by the Comet assay (Figure 5A-B).
Figure 5.
Accumulation of single- or double-stranded DNA breaks. (A) Alkaline single cell gel electrophoresis (Comet assay) of CD4+CD8− (ISP) and CD4+CD8+ (DP) subpopulations for control, SCID P1, P2, and OS P3 cells. DNA was visualized by SYBR Gold. The scale bar corresponds to 100 μm. Tail lengths were measured and summarized in (B). ****P < .0001; **P < .01.
V(D)J gene recombination analysis of early T cells from OS- and SCID-derived iPSC lines
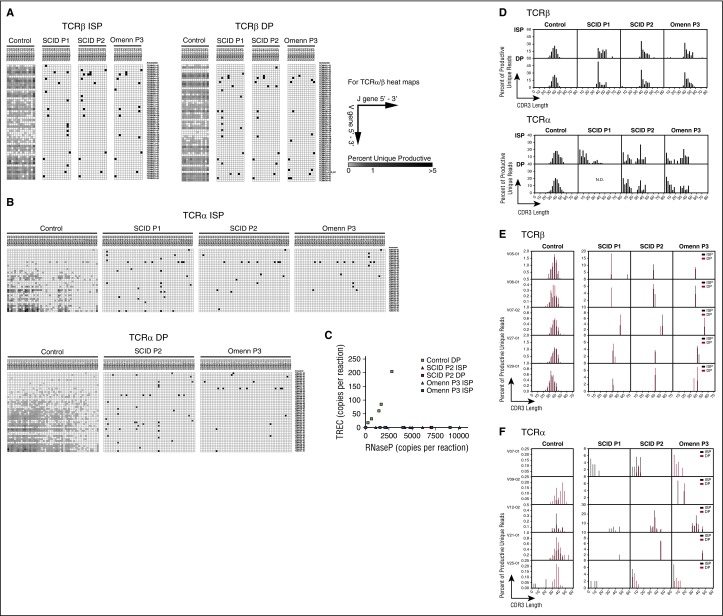
In spite of severe lymphoid depletion in the thymus,30 patients with OS have a variable number of circulating T cells, which however, lack a diverse TCR repertoire. To determine to what extent restriction of the TCR repertoire is already present at early stages of T-cell development in patients with OS, deep sequencing analysis of the TRB and TRA V(D)J rearrangements was performed on genomic DNA extracted from sorted CD4+ ISP and CD4+ CD8+ DP cells generated in vitro from control-, OS-, and SCID-derived iPSCs (Table 1). Due to low cell numbers, the SCID P1 DP population was not analyzed for TRA rearrangement. A reduction in the proportion of unique and total TRB and TRA rearranged sequences was evident in OS and SCID samples as compared with control (supplemental Figure 5A). Broad diversity of TRBV and TRAV gene usage and of V-J pairing was demonstrated among both unique (Figure 6A-B) and total (supplemental Figure 5B-C) sequences from sorted ISP and DP cells derived from control iPSCs, whereas a dramatic restriction of TRB and TRA repertoires was observed in equivalent OS and SCID cell subsets (Figure 6A-B; supplemental Figure 5B-C). To further analyze the ability of control- and RAG1-mutated cells to accomplish rearrangements at the TRA/δ locus, we quantified the levels of TRECs in sorted DP cells. TRECs were readily detectable in control, but not in SCID or OS cells (Figure 6C). Virtual spectratyping analysis of the distribution of TRB CDR3 length among unique sequences showed a normal distribution for ISP and DP cells derived from control iPSCs, whereas OS and SCID cells were characterized by a skewed distribution of CDR3 lengths, with presence of longer sequences (Figure 6D, top panel). Similarly, a skewed distribution of CDR3 lengths was also observed when analyzing the most abundant V-J TRB rearrangements (Figure 6E). Interestingly, TRA CDR3 length distribution revealed two peaks in SCID and OS cells, suggesting that some clones have normal CDR3 length whereas others exhibit an abnormally reduced length (Figure 6D, lower panel). Indeed, when analyzing some of the most abundant V-J TRA rearrangements, both shortened (V07-01 and V25-01) or normal (V12-02 and V21-01) CDR3 lengths can be seen (Figure 6F).
Table 1.
Summary of deep sequencing analysis of the TRB V(D)J rearrangements
| Cell line | Population | Total | Unique | Productive total | Productive unique | Clonality |
|---|---|---|---|---|---|---|
| Control | CD4+ CD8− | 451 195 | 3 300 | 428 110 | 2512 | 0.52 |
| 603 206 | 6 949 | 478 446 | 2875 | 0.52 | ||
| Control | CD4+ CD8+ | 734 020 | 8 775 | 666 299 | 6930 | 0.30 |
| 900 284 | 14 151 | 724 386 | 8414 | 0.28 | ||
| SCID P1 | CD4+ CD8− | 1 026 133 | 366 | 1 021 305 | 219 | 0.60 |
| Omenn P3 | CD4+ CD8− | 132 386 | 120 | 131 048 | 76 | 0.49 |
| Omenn P3 | CD4+ CD8+ | 784 927 | 301 | 782 376 | 217 | 0.65 |
Figure 6.
Next generation sequencing analysis of TCR repertoire upon in vitro T-lineage differentiation of control, SCID (P1 and P2), and OS (P3)-derived iPSCs. (A) Heat map representation of percentage of TRB VJ (orientated via chromosomal 5′ to 3′ distribution) pairings among unique sequences in ISP (left) and DP (right) T-lineage cells derived from the indicated patient iPS lines. Results demonstrate 1 representative sample from 2 experiments with similar results. (B) Heat map representation of percentage of TRA VJ (orientated via chromosomal 5′ to 3′ distribution) pairings among total sequences (all δ rearrangements excluded from analysis) in ISP (left) and DP (right) T-lineage cells derived from the indicated patients’ iPS lines among unique sequences. Results demonstrate 1 representative sample from 2 experiments with similar results. (C) Quantitative PCR analysis of TRECs in control, SCID P2, and OS P3 cells. RNase P was used as an internal control for quality of genomic DNA amplification. (D) Virtual spectratyping, showing skewing in the distribution of CDR3 lengths among unique TRB or TRA sequences expressed by ISP (top) and DP (bottom) cells in SCID P1, P2, and Omenn P3 compared with control. (E) Distribution of CDR3 length for 5 more commonly expressed V genes in each sample for unique TRB sequences. (F) Distribution of CDR3 length for 5 more commonly expressed V genes in each sample for unique TRA sequences. Results demonstrate 1 representative sample from 2 experiments with similar results. RNase P, ribonuclease P.
Finally, tree map representation of CDR3 amino acid sequences showed increased diversity, and reduced clonotypic expansion, in ISP and DP cells from control as compared with SCID and OS cells for both TRB (supplemental Figure 5E) and TRA (supplemental Figure 5F) repertoires.
Discussion
Combined immunodeficiencies include a heterogeneous group of conditions, whose broad clinical and immunologic phenotype depends on the nature of the gene affected, and the specific mutations involved.31,32 Here, we initially observed normal expression of markers corresponding to early stages of T-cell development (CD7, CD5, and CD38), which was also seen in cells with RAG1 mutations associated with SCID and OS. However, these cells showed delayed expression of CD5 and CD38, resulting in a partial block at the CD7+CD5−CD38− stage. Nonetheless, both SCID- and OS-derived cells maintained some ability to initially express CD4, CD8α, and CD8β. Prior observations in the mouse that RAG1/2 expression occurs at the CD4−CD8− double-negative stage, but is downregulated by pre-TCR signal to allow maturation to the CD4+CD8+ DP stage, followed by a second wave of RAG expression and TRA recombination.33 The emergence of a low number of DP cells during culture of iPSC-derived CD34+ cells from patients with RAG1-deficient SCID suggests that the first wave of RAG1 expression and V(D)J recombination are not strictly required for the development of DP cells in humans, or that even these seemingly null RAG1 mutations support minimal levels of recombination activity that suffice for partial progression to the DP stage. Control cells continued to mature, as demonstrated by expression of CD3 and TRA/TRB, whereas the presence of DP cells was rapidly lost in culture of SCID- and OS-derived cells, where only CD5+CD7hi cells were maintained. At later time points, SCID- and OS-derived cells remained as CD38−, CD31−, and CD45RA+, which is consistent with a failure to effectively traverse the pre–TCR-dependent β-selection checkpoint of T-cell differentiation.
Although the RAG1 mutation associated with OS supported residual levels of recombination activity when tested with the Abelson pro–B-cell system, iPSC-derived CD34+ cells from the patient with OS did not show an enhanced ability to support T-cell differentiation in vitro when compared with SCID-derived cells. It is possible that the residual RAG1 function in OS-derived cells is sufficient to initiate V(D)J recombination, but that these cells fail to survive and cannot progress to later stages. RAG1 protein plays an indispensable role in DNA nicking and cutting, and it is also involved in the joining process. Southern blot analysis revealed that introduction of the hypomorphic c.256_257delAA RAG1 mutant into Abelson-transformed pro-B cells did not result in accumulation of cells with unresolved DNA-DSBs, as observed for ligase IV-deficient cells. These data are consistent with the notion that this RAG1 mutation associated with OS is defective in one of the steps prior to double-strand DNA cleavage. The assay is limited by the requirement of a specific probe, therefore is unable to detect promiscuous, off-target DNA-DSBs that could conceivably occur in cells with mutant RAG1, nor does it show single-strand breaks. To address both possibilities, we further analyzed the efficiency of DNA cleavage of RAG1 mutants associated with OS by employing the single cell gel electrophoresis (Comet assay), which was performed under alkaline conditions in order to reveal single- and double-strand DNA breaks. Surprisingly, CD4+CD8− and CD4+CD8+ derived from the patient with OS consistently yielded comets. One possible explanation is that the hypomorphic RAG1 mutant may inappropriately nick DNA, while failing to introduce appropriate DNA-DSBs. According to this idea, the initial hydrolysis step of cleavage would be functional, but the reaction would be selectively defective for the subsequent transesterification step of hairpin formation, in which the 3′ hydroxyl freed by single-strand cleavage attacks the backbone of the other strand. Single-strand cuts (or off-target DNA-DSBs) could accumulate and eventually lead to cell death, whereas on-site DNA-DSBs would yield the observed low-level and skewed recombination outcomes. To test the SCID and OS mutants directly for nicking activity, we used the catalytic domain of SCID P2 and OS P3 to conduct an oligonucleotide cleavage assay, which revealed that these mutants have no nicking activity nor can they bind DNA (supplemental Figure 4A-B). Although the N-terminus truncated mutant of OS P3 retains an intact catalytic core, it is predominantly retained in the cytoplasm, and therefore has severely restricted activity.26 However, OS P3 was a compound heterozygote for the c.256_257delAA and the c.2164G>A (p.E722K) mutations. To explain the presence of comets and the inability to complete T-cell differentiation in OS P3, we have developed a model (supplemental Figure 6) that takes into account that dimerization of RAG1 precedes nuclear translocation of the RAG complex. Swanson et al have shown that a heterodimer containing cleavage-competent and cleavage-incompetent components can nick DNA efficiently.34 Similarly, a heterodimer of the RAG1 mutants in OS P3 cells could enter the nucleus via E722K and bind DNA through the N-terminus truncated protein, which can also nick one RSS. However, the second site cannot be nicked by E722K, which has no activity, thereby preventing hairpin formation.35,36 The RAG1 remains tightly bound to the site of the nick,37 and cells with single-strand DNA breaks accumulate and are readily observed by the Comet assay. However, persistence of single-strand DNA breaks in immature T-lineage cells would culminate in impaired cell survival, and thus failure to sustain T-cell differentiation.
A recent publication by Teng et al demonstrated that, although RAG1 can bind ∼1800 sites in human thymocytes, off-target activity is prevented by reducing cryptic recombination signals near RAG1 binding sites,38 thereby protecting the genome against oncogenesis. In light of this, our data indicate that DNA nicks or breaks are largely resolved when WT RAG1 is present, or cannot occur in SCID P1 or P3 cells with catalytically inactive RAG1, whereas these breaks persist in OS P3 cells (Figure 5). Assessing whether some RAG1 mutations result in recognition of different off-target sites would be important, as the inherent protective mechanisms would not be in place, thereby increasing the risk of oncogenic genome rearrangements.
The observation that in vitro T-cell differentiation was similarly inefficient, both for SCID and for the OS patient, is consistent with the notion that OS-associated RAG mutations are very severe and support minimal levels of DNA recombination. This was also reflected by the severe restriction of TRB and TRA rearrangements that was documented in sorted ISP and DP cells from patients with SCID and OS as compared with control cells. These data also suggest that the oligoclonality of peripheral T cells that has been reported in patients with OS39,40 is not solely due to peripheral selection, but also reflects an intrinsic and severe defect of V(D)J recombination that occurs in the thymus, as also indicated by previous studies from our group in post-mortem thymic samples from patients with OS.30 Control cells showed a clear bias for proximal Va (3′-end) and Ja (5′-end) usage, whereas remarkably this was not the case for RAG1-mutant cells. To date, this is the first comparison of both TRB and TRA rearrangement of in vitro-derived SCID and OS T-lineage cells, and further studies are required to elucidate the causality for the observed lack of proximal bias observed for TRA rearrangements in RAG1-deficient cells.
In addition to the overall restriction of TRB and TRA rearrangements, in vitro differentiated ISP and DP cells from patients with SCID or OS revealed a series of additional abnormalities, including preferential usage of certain V and J genes, and skewing of CDR3 length, with presence of unusually long or short CDR3 fragments. Similar abnormalities have been recently reported in peripheral T cells from patients with OS.41 Variability of the length of the CDR3 loop is determined by V(D)J recombination.42 Although CDR3 lengths are typically highly constrained for the TRA/TRB chains,43 a slight decrease in CDR3 length is observed as T lymphocytes mature from the ISP to the DP stage.44 To this end, the increased TRB CDR3 length that we have observed in sorted ISP and DP cells from patients with SCID and OS may indicate that these cells express a less mature TCR that may escape central tolerance, thus allowing autoreactive T cells to exit the thymus and contribute to the OS phenotype.
In summary, we have demonstrated that iPSCs represent a valuable tool to study mechanisms of altered T-cell development in patients with SCID and OS, and may offer unique insights into genotype-phenotype correlation in patients carrying distinct mutations in the same gene.
Acknowledgments
The authors thank their patients, their families, and physicians; and the Courtney McIntosh and Vincent Cheng (Centre for Flow Cytometry and Microscopy, Sunnybrook Research Institute, Toronto, ON, Canada) for their expert cell sorting support, and Andrew Elias (Princess Margaret Cancer Centre, Toronto, ON, Canada) for his assistance with fluorescence microscopy.
This work was supported by grants from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (1R01AI100887 [L.D.N. and J.C.Z.-P.] and 2U54AI082973 [L.D.N.]), the March of Dimes (1-FY13-500) (L.D.N.), and by The Krembil Foundation (J.C.Z.-P). J.C.Z.-P. is also supported by a Canada Research Chair in Developmental Immunology. P.M.B. was supported by a Canadian Institutes of Health Research Postdoctoral Fellowship award. I.M.P. is supported by the Israeli Centers of Research Excellence, Gene Regulation in Complex Human Disease, Center No 41/11. F.D.B. and E.C. were supported in part by R01AI 082020.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.M.B., L.D.N., and J.C.Z.-P. designed research; P.M.B., I.M.P., E.C., L.O.d.B., Y.N.L., C.D.-B., J.H.R., A.M.C., G.A., K.F., Y.H.Z., A.B., L.D., and F.V. performed experiments; S.G., M.K., G.K., B.P.S., W.A.-H., D.G.S., and F.D.B. contributed reagents/training; P.M.B., E.C., D.G.S., F.D.B., L.D.N., and J.C.Z.-P. analyzed data; and P.M.B., L.D.N., and J.C.Z.-P. prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Luigi D. Notarangelo, Division of Immunology, Boston Children’s Hospital, 1 Blackfan Cir, Karp Research Building, Room 10217, Boston, MA 02115; e-mail: luigi.notarangelo@childrens.harvard.edu; and Juan Carlos Zúñiga-Pflücker, Sunnybrook Research Institute, 2075 Bayview Ave, Room A-331, Toronto, ON M4N 3M5, Canada; e-mail: jczp@sri.utoronto.ca.
References
- 1.Picard C, Al-Herz W, Bousfiha A, et al. Primary immunodeficiency diseases: an pdate on the classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. J Clin Immunol. 2015;35(8):696–726. doi: 10.1007/s10875-015-0201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(suppl):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 3.Roth DB, Menetski JP, Nakajima PB, Bosma MJ, Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70(6):983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 4.Fugmann SD. RAG1 and RAG2 in V(D)J recombination and transposition. Immunol Res. 2001;23(1):23–39. doi: 10.1385/IR:23:1:23. [DOI] [PubMed] [Google Scholar]
- 5.Notarangelo LD, Kim MS, Walter JE, Lee YN. Human RAG mutations: biochemistry and clinical implications. Nat Rev Immunol. 2016;16(4):234–246. doi: 10.1038/nri.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villa A, Santagata S, Bozzi F, et al. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93(5):885–896. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- 7.Giblin W, Chatterji M, Westfield G, et al. Leaky severe combined immunodeficiency and aberrant DNA rearrangements due to a hypomorphic RAG1 mutation. Blood. 2009;113(13):2965–2975. doi: 10.1182/blood-2008-07-165167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuetz C, Huck K, Gudowius S, et al. An immunodeficiency disease with RAG mutations and granulomas. N Engl J Med. 2008;358(19):2030–2038. doi: 10.1056/NEJMoa073966. [DOI] [PubMed] [Google Scholar]
- 9.Abolhassani H, Wang N, Aghamohammadi A, et al. A hypomorphic recombination-activating gene 1 (RAG1) mutation resulting in a phenotype resembling common variable immunodeficiency. J Allergy Clin Immunol. 2014;134(6):1375–1380. doi: 10.1016/j.jaci.2014.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YN, Frugoni F, Dobbs K, et al. A systematic analysis of recombination activity and genotype-phenotype correlation in human recombination-activating gene 1 deficiency. J Allergy Clin Immunol. 2014;133(4):1099–1108. doi: 10.1016/j.jaci.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosma GC, Custer RP, Bosma MJ. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301(5900):527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- 12.Blunt T, Finnie NJ, Taccioli GE, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80(5):813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 13.Bosma MJ, Carroll AM. The SCID mouse mutant: definition, characterization, and potential uses. Annu Rev Immunol. 1991;9:323–350. doi: 10.1146/annurev.iy.09.040191.001543. [DOI] [PubMed] [Google Scholar]
- 14.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68(5):869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 15.Shinkai Y, Rathbun G, Lam KP, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68(5):855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 16.Khiong K, Murakami M, Kitabayashi C, et al. Homeostatically proliferating CD4 T cells are involved in the pathogenesis of an Omenn syndrome murine model. J Clin Invest. 2007;117(5):1270–1281. doi: 10.1172/JCI30513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marrella V, Poliani PL, Casati A, et al. A hypomorphic R229Q Rag2 mouse mutant recapitulates human Omenn syndrome. J Clin Invest. 2007;117(5):1260–1269. doi: 10.1172/JCI30928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perryman LE. Molecular pathology of severe combined immunodeficiency in mice, horses, and dogs. Vet Pathol. 2004;41(2):95–100. doi: 10.1354/vp.41-2-95. [DOI] [PubMed] [Google Scholar]
- 19.Wiler R, Leber R, Moore BB, VanDyk LF, Perryman LE, Meek K. Equine severe combined immunodeficiency: a defect in V(D)J recombination and DNA-dependent protein kinase activity. Proc Natl Acad Sci USA. 1995;92(25):11485–11489. doi: 10.1073/pnas.92.25.11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spits H. Development of alphabeta T cells in the human thymus. Nat Rev Immunol. 2002;2(10):760–772. doi: 10.1038/nri913. [DOI] [PubMed] [Google Scholar]
- 21.Kennedy M, Awong G, Sturgeon CM, et al. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Reports. 2012;2(6):1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Vizcardo R, Masuda K, Yamada D, et al. Regeneration of human tumor antigen-specific T cells from iPSCs derived from mature CD8(+) T cells. Cell Stem Cell. 2013;12(1):31–36. doi: 10.1016/j.stem.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Chang CW, Lai YS, Lamb LS, Jr, Townes TM. Broad T-cell receptor repertoire in T-lymphocytes derived from human induced pluripotent stem cells. PLoS One. 2014;9(5):e97335. doi: 10.1371/journal.pone.0097335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menon T, Firth AL, Scripture-Adams DD, et al. Lymphoid regeneration from gene-corrected SCID-X1 subject-derived iPSCs. Cell Stem Cell. 2015;16(4):367–372. doi: 10.1016/j.stem.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shearer WT, Dunn E, Notarangelo LD, et al. Establishing diagnostic criteria for severe combined immunodeficiency disease (SCID), leaky SCID, and Omenn syndrome: the Primary Immune Deficiency Treatment Consortium experience. J Allergy Clin Immunol. 2014;133(4):1092–1098. doi: 10.1016/j.jaci.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santagata S, Gomez CA, Sobacchi C, et al. N-terminal RAG1 frameshift mutations in Omenn’s syndrome: internal methionine usage leads to partial V(D)J recombination activity and reveals a fundamental role in vivo for the N-terminal domains. Proc Natl Acad Sci USA. 2000;97(26):14572–14577. doi: 10.1073/pnas.97.26.14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pessach IM, Ordovas-Montanes J, Zhang SY, et al. Induced pluripotent stem cells: a novel frontier in the study of human primary immunodeficiencies. J Allergy Clin Immunol. 2011;127(6):1400–1407. doi: 10.1016/j.jaci.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinacht KG, Brauer PM, Felgentreff K, et al. The role of induced pluripotent stem cells in research and therapy of primary immunodeficiencies. Curr Opin Immunol. 2012;24(5):617–624. doi: 10.1016/j.coi.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun. 1984;123(1):291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- 30.Signorini S, Imberti L, Pirovano S, et al. Intrathymic restriction and peripheral expansion of the T-cell repertoire in Omenn syndrome. Blood. 1999;94(10):3468–3478. [PubMed] [Google Scholar]
- 31.Milner JD, Holland SM. The cup runneth over: lessons from the ever-expanding pool of primary immunodeficiency diseases. Nat Rev Immunol. 2013;13(9):635–648. doi: 10.1038/nri3493. [DOI] [PubMed] [Google Scholar]
- 32.Villa A, Sobacchi C, Notarangelo LD, et al. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97(1):81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 33.Wilson A, Held W, MacDonald HR. Two waves of recombinase gene expression in developing thymocytes. J Exp Med. 1994;179(4):1355–1360. doi: 10.1084/jem.179.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swanson PC. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol Cell Biol. 2001;21(2):449–458. doi: 10.1128/MCB.21.2.449-458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu K, Lieber MR. Mechanistic basis for coding end sequence effects in the initiation of V(D)J recombination. Mol Cell Biol. 1999;19(12):8094–8102. doi: 10.1128/mcb.19.12.8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bischerour J, Lu C, Roth DB, Chalmers R. Base flipping in V(D)J recombination: insights into the mechanism of hairpin formation, the 12/23 rule, and the coordination of double-strand breaks. Mol Cell Biol. 2009;29(21):5889–5899. doi: 10.1128/MCB.00187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grawunder U, Lieber MR. A complex of RAG-1 and RAG-2 proteins persists on DNA after single-strand cleavage at V(D)J recombination signal sequences. Nucleic Acids Res. 1997;25(7):1375–1382. doi: 10.1093/nar/25.7.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng G, Maman Y, Resch W, et al. RAG represents a widespread threat to the lymphocyte genome. Cell. 2015;162(4):751–765. doi: 10.1016/j.cell.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rieux-Laucat F, Bahadoran P, Brousse N, et al. Highly restricted human T cell repertoire in peripheral blood and tissue-infiltrating lymphocytes in Omenn’s syndrome. J Clin Invest. 1998;102(2):312–321. doi: 10.1172/JCI332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brooks EG, Filipovich AH, Padgett JW, Mamlock R, Goldblum RM. T-cell receptor analysis in Omenn’s syndrome: evidence for defects in gene rearrangement and assembly. Blood. 1999;93(1):242–250. [PubMed] [Google Scholar]
- 41.Yu X, Almeida JR, Darko S, et al. Human syndromes of immunodeficiency and dysregulation are characterized by distinct defects in T-cell receptor repertoire development. J Allergy Clin Immunol. 2014;133(4):1109–1115. doi: 10.1016/j.jaci.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes MM, Yassai M, Sedy JR, et al. T cell receptor CDR3 loop length repertoire is determined primarily by features of the V(D)J recombination reaction. Eur J Immunol. 2003;33(6):1568–1575. doi: 10.1002/eji.200323961. [DOI] [PubMed] [Google Scholar]
- 43.Rock EP, Sibbald PR, Davis MM, Chien YH. CDR3 length in antigen-specific immune receptors. J Exp Med. 1994;179(1):323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishio J, Suzuki M, Nanki T, Miyasaka N, Kohsaka H. Development of TCRB CDR3 length repertoire of human T lymphocytes. Int Immunol. 2004;16(3):423–431. doi: 10.1093/intimm/dxh046. [DOI] [PubMed] [Google Scholar]