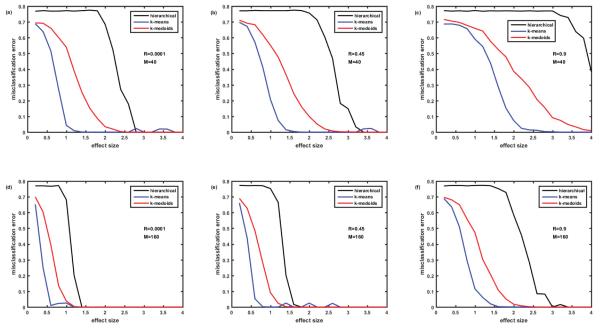
Figure 6.
Comparison of misclassification error rates generated by three clustering algorithms: hierarchical, k-means, and k-medoids. Cohort of 100 simulated patients (5 clusters of equal size). Protein assay includes 1129 target proteins. Protein abundances are correlated ‘among neighbors’ - Rij=R∣i-j∣. Case of completely overlapping biomarker signatures. Signatures of the clusters (disease subtypes) differ by the signs of the effect (up- or down-regulation of the proteins). See details in the text. Figures 6A-6C- M=40 biomarkers in the signature. Figures 6D-6F– M=160 biomarkers in the signature. Figures 6A and 6D– correlation coefficient R=0.0001, Figures 6B and 6E– R=0.45, Figures 6C and 6F– R=0.9.