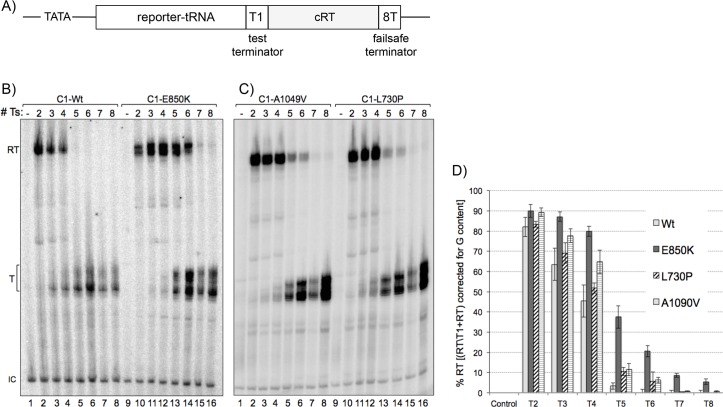
Fig 5. Promoter-dependent in vitro transcription reveals oligo(T) length-dependent terminator readthrough by RNAP III C1 mutants.
A) Schematic of tRNA gene arrangement used for in vitro transcription by S100 extracts; the plasmids used for transcription differed only in the number of Ts at the test terminator, T1, as listed above the lanes of B & C. B-C) S100 extracts from yKR1-C1 mutants or -C1-Wt control were used as a source of initiation factors and RNAP III as indicated above the lanes. Plasmids containing tRNA genes that differ only in the number of Ts in the oligo(T) terminator as indicated above the lanes were used as templates to program the transcription reactions. RT = readthrough; termination at the failsafe terminator, T = termination at the test terminator, IC = internal control. D) Quantitation of % readthrough as defined on the Y-axis; reactions contained 32P-αGTP.