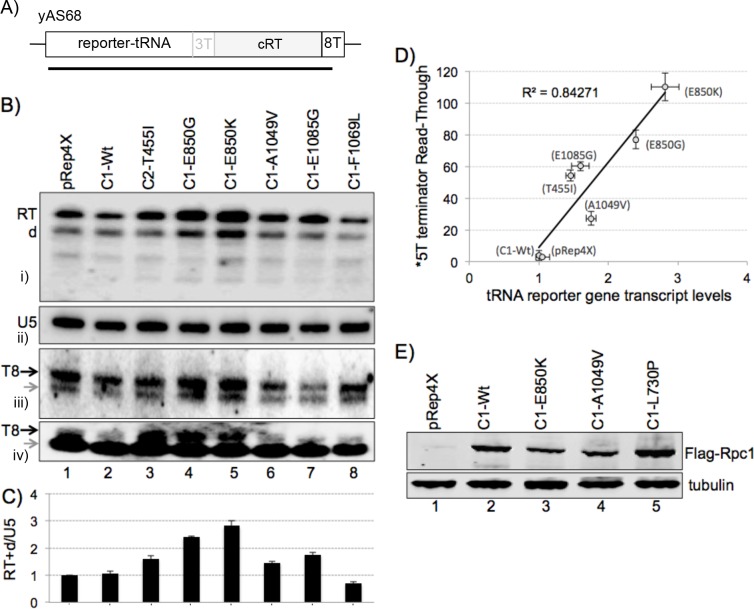
Fig 9. RNAP III C1 mutants increase RNA production from a tRNA gene.
A) Schematic of the tRNA reporter gene in the strain yAS68 and the RT transcript produced from the 8T terminator. B) tRNA reporter gene readthrough (RT) transcript-specific probing of RNA from C1-mutants and controls as indicated above the lanes. Transcription of this tRNA reporter gene makes only RT transcripts and not the normal pre-tRNAs (upper panel, i, see text). The second panel (ii) shows probing of the same blot as in upper panel, for U5 snRNA. The next lower panel (iii) shows probing for the 3'-trailer of a nascent pre-tRNASerGCU whose endogenous single copy gene bears an 8T terminator (black arrow); the small grey arrow points to an intron-containing, 3'-trailer-containing intermediate. The lowest panel (iv) shows probing for the unique 3' trailer of a single copy nascent pre-tRNAValUAC whose gene does not contain an intron and ends with a T8 terminator (Black arrow); the small grey arrow points to the more abundant mature tRNAValUAC product of two genes that overlap with this probe. C) Quantification of data as in (A) from duplicate experiments calibrated by U5 RNA on the same blots; note that C1-F1069L, lane 8, is from a GOF mutant. D) RT transcript (Y-axis) from 5T terminator RT data plotted against amount of tRNA reporter gene output (X-axis) as in panel A; each represented by duplicate data sets reflected by error bars. The Y-axis represents a proxy for relative elongation rate (see text). E) Cells transformed with the pRep4X vector, C1-mutant or C1-Wt alleles carrying a Flag tag were subjected to immunoblotting for their Rpc1 protein.