Abstract
Background
Succinate has been identified by the U.S. Department of Energy as one of the top 12 building block chemicals, which can be used as a specialty chemical in the agricultural, food, and pharmaceutical industries. Escherichia coli are now one of the most important succinate producing candidates. However, the stoichiometric maximum succinate yield under anaerobic conditions through the reductive branch of the TCA cycle is restricted by NADH supply in E. coli.
Results
In the present work, we report a rational approach to increase succinate yield by regulating NADH supply via pentose phosphate (PP) pathway and enhancing flux towards succinate. The deregulated genes zwf243 (encoding glucose-6-phosphate dehydrogenase) and gnd361 (encoding 6-phosphogluconate dehydrogenase) involved in NADPH generation from Corynebacterium glutamicum were firstly introduced into E. coli for succinate production. Co-expression of beneficial mutated dehydrogenases, which removed feedback inhibition in the oxidative part of the PP pathway, increased succinate yield from 1.01 to 1.16 mol/mol glucose. Three critical genes, pgl (encoding 6-phosphogluconolactonase), tktA (encoding transketolase) and talB (encoding transaldolase) were then overexpressed to redirect more carbon flux towards PP pathway and further improved succinate yield to 1.21 mol/mol glucose. Furthermore, introducing Actinobacillus succinogenes pepck (encoding phosphoenolpyruvate carboxykinase) together with overexpressing sthA (encoding soluble transhydrogenase), further increased succinate yield to 1.31 mol/mol glucose. In addition, removing byproduct formation through inactivating acetate formation genes ackA-pta and heterogenously expressing pyc (encoding pyruvate carboxylase) from C. glutamicum led to improved succinate yield to 1.4 mol/mol glucose. Finally, synchronously overexpressing dcuB and dcuC encoding succinate exporters enhanced succinate yield to 1.54 mol/mol glucose, representing 52 % increase relative to the parent strain and amounting to 90 % of the strain-specific stoichiometric maximum (1.714 mol/mol glucose).
Conclusions
It’s the first time to rationally regulate pentose phosphate pathway to improve NADH supply for succinate synthesis in E. coli. 90 % of stoichiometric maximum succinate yield was achieved by combining further flux increase towards succinate and engineering its export. Regulation of NADH supply via PP pathway is therefore recommended for the production of products that are NADH-demanding in E. coli.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-016-0536-1) contains supplementary material, which is available to authorized users.
Keywords: Escherichia coli, Succinate, Anaerobic, Pentose phosphate pathway, Transhydrogenase, Cofactor
Background
Succinate and its derivatives are widely used as specialty chemicals in the field of food, pharmaceutical, and cosmetic industries [1]. The industrial potential for succinate fermentations was recognized as early as 1980 [2]. Several bacteria, such as, Anaerobiospirillum succiniciproducens [3], Actinobacillus succinogenes [4], and Mannheimia succiniciproducens [5], have been isolated and manipulated for succinate production. However, those rumen organisms usually require high-cost nutrient sources for growth and perform characteristically unstable in the process of fermentation. Recent studies on metabolic engineering of Escherichia coli for succinate production have been carried out due to the ease of genetic manipulation coupled to its rapid growth rate, standardized cultivation techniques, and inexpensive media.
In anaerobic conditions, E. coli proceeds with mixed-acid fermentation but succinate represents a minor product [6]. Derivatives of E. coli have been constructed in efforts to improve succinate production with variable success. Most genetic engineering strategies focused on eliminating competing pathways [7–9], overexpressing native or heterologous key enzymes [9–12], disrupting phosphotransferase (PTS) system to improve PEP supply [12, 13], activating glyoxylate pathway [14, 15], and combining metabolic engineering and metabolic evolution [11, 16, 17]. Some other external means, such as a dual phase fermentation production mode which comprises an initial aerobic growth phase followed by an anaerobic production phase, have also been developed in order to increase succinate production [18].
In wild type E. coli, the reductive tricarboxylic acid (TCA) branch significantly contributed to anaerobic succinate production (Fig. 1). One major obstacle to high succinate yield through this fermentative pathway is NADH availability [14, 17]. Inactivating NADH competing pathways has often been applied to improve NADH availability for succinate production [19], while deletion of these genes that block the competing routes for NADH oxidation were insufficient to direct carbon flow to succinate [20]. Glycerol, as an alternative substrate to glucose, has become an appealing substrate for biological synthesis and was applied to succinate synthesis due to its high degree of reduction [21]. When glycerol was used as a carbon source in the anaerobic fermentation of E. coli, higher levels of NADH are generated in glycerol dissimilation through glycerol dehydrogenase (GldA) [21, 22]. Recently, the NAD+-dependent formate dehydrogenase from Candida boidinii was introduced into an E. coli strain [23]. The introduced formate dehydrogenase converted formate into NADH and CO2, which not only increased succinate productivity but reduced concentrations of byproduct formate. However, this strategy is usually associated with the carbon source loss or requires formate addition. In addition, the glyoxylate pathway can also form succinate, which is essentially active under aerobic condition, and conserves NADH consumption relative to reductive TCA branch in succinate formation [14, 15]. It has been reported that succinate yield can reach its maximum level 1.714 mol/mol glucose by redirecting 71.4 % of carbon flows to the reductive TCA branch and 28.6 % of the carbon flows to the glyoxylate pathway [10], while employment of the glyoxylate bypass for succinate synthesis is accompanied by the CO2 loss [24].
Fig. 1.
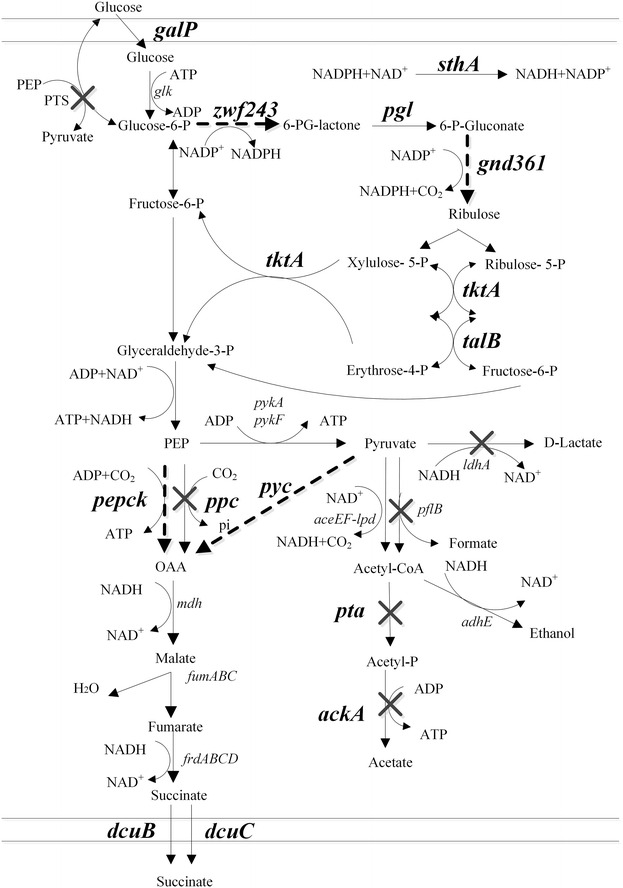
Metabolic network of E. coli under anaerobic conditions and metabolic engineering strategies for succinate overproduction. Modified genes for improving succinate yield are highlighted in bold. X indicates metabolic reactions that have been blocked by gene deletions. Dotted arrows indicate steps that are heterogeneously expressed in E. coli. Genes and enzymes: ldhA encoding lactate dehydrogenase; pflB encoding pyruvate-formate lyase; pta encoding phosphate acetyltransferase; ackA encoding acetate kinase; adhE encoding alcohol dehydrogenase; ppc encoding phosphoenolpyruvate carboxylase; pepck encoding phosphoenolpyruvate carboxykinase; pyc encoding pyruvate carboxylase; aceEF-lpd encoding pyruvate dehydrogenase complex; mdh encoding malate dehydrogenase; fumA, fumB, and fumC encoding fumarase isozymes; frdABCD encoding fumarate reductase; galp encoding galactose permease; glk encoding glucokinase; PTS phosphotransferase systems; zwf243 encoding 6-phosphate dehydrogenase; gnd361 encoding 6-phosphogluconate dehydrogenase; pgl encoding 6-phosphogluconolactonase; tktA encoding transketolase; talB encoding transaldolase; sthA encoding soluble transhydrogenase
Pentose phosphate (PP) pathway was known to play major roles in NADPH generation and modulation of the PP pathway has been conventionally attempted as primary solution to increase NADPH-dependent biotransformation [25, 26]. In E. coli, there exist two types of pyridine nucleotide transhydrogenase which are responsible for the mutual transformation between NADPH and NADH [26, 27]. They are energy-dependent membrane-bound transhydrogenase (TH or PntAB, EC 1.6.1.2) and energy-independent soluble transhydrogenase (SthA or UdhA, EC 1.6.1.1). The membrane-bound PntAB catalyzes the transfer of a hydrogen ion from NADH to NADPH under low levels of NADPH, while the soluble SthA catalyzes the transfer of a hydrogen ion from NADPH to NADH under high concentrations of NADPH [26]. Recently, a high-succinate-producing strain HX024 was obtained through metabolic evolution. It was found that pyruvate dehydrogenase (PDH) was significantly activated, and sensitivity of PDH to NADH was eliminated by three mutations in LpdA, which is the E3 component of PDH. Another reasons for high yield of strain HX024 were ascribed to the increased transketolase and soluble transhydrogenase SthA activities [17]. It should be noted that PP pathway converts glucose-6-phosphate into fructose-6-phosphate and glyceraldehyde-3-phosphate with NADPH produced, the carbon flux from synthesized fructose-6-phosphate and glyceraldehyde-3-phosphate then go through glycolytic pathway and produce PEP and NADH, and NADPH could be converted into NADH by native transhydrogenase SthA for succinate synthesis. Therefore, it should be a potential strategy to regulate NADH supply for succinate synthesis by rationally modifying PP pathway in E. coli.
In the present work, a reducing equivalent-conserving pathway, which combined the coordinated re-engineering of the glycolytic pathway and the pentose phosphate pathway, was implemented for the first time to provide more NADH to achieve the stoichiometric maximum succinate yield 1.714 mol/mol glucose. NADH availability was then increased for succinate synthesis through modifying pentose phosphate pathway. Systems metabolic engineering was followed to further improve succinate yield, which involves in increasing carbon flux towards succinate, reinforcing NADH level, and finally engineering succinate export. As a result of these modifications, 90 % of stoichiometric maximum succinate yield was achieved. The strategy for improving NADH supply described here may be useful for engineering E. coli strains for the industrial production of succinate and related NADH-demanding products.
Results
Rationale for enhancing NADH supply by modifying pentose phosphate pathway
We presumed that succinate is formed exclusively via the reductive TCA branch. Glucose is internalized into the cytoplasm and then catalyzed into glucose-6P by glucokinase (encoded by glk), which was then catabolized into PEP and NADH for succinate synthesis. The cells are capable of producing PEP and NADH from glucose at two different modes.
The first mode corresponds to the formation of PEP and NADH exclusively via glycolytic pathway, the reaction may be expressed as:
| 1 |
The second mode implies the involvement of pentose phosphate pathway in the PEP and NADH formation, the reaction may be expressed as:
| 2 |
NADPH could be converted into NADH by transhydrogenase SthA, the reaction may be expressed as:
| 3 |
Integrating of the reaction (2), (3), we can get the reaction:
| 4 |
PEP was reduced to become succinate via reductive TCA branch, in which two molar NADH were required. The reaction may be expressed as:
| 5 |
It is not difficult to find that NADH is in excess for reducing PEP to from succinate via PP pathway in reaction (4), while NADH is insufficient for reducing PEP to from succinate in reaction (1). According to redox balance, the maximum stoichiometric succinate yield of 1.714 mol/mol glucose can be obtained if the carbon flux ratio between PP pathway and glycolysis pathway is 6:1.
Succinate yield is effectively enhanced by redirection of flux towards pentose phosphate pathway
As shown in Fig. 1, glucose-6-phosphate dehydrogenase (encoded by zwf) and 6-phosphogluconate dehydrogenase (encoded by gnd) are involved in NADPH generation in the oxidative part of the PP pathway, and many studies have focused on the modulation of these two endogenous enzymes to improve NADPH-dependent biotransformation in E. coli [26]. While both enzymes are inhibited by NADPH, ATP, fructose 1,6-bisphosphate (FBP) and the 6PGD additionally by D-glyceraldehyde 3-phosphate (Gra3P), erythrose 4-phosphate and Ru5P. The feedback regulation implies that PP pathway can respond very flexibly to altered NADPH and product levels. Overexpression of the deregulated genes zwf243 (with point mutation A243T) and gnd361 (with point mutation S361F), which led to release of feedback inhibition, significantly redirected carbon flow through the PP pathway and increased lysine production in Corynebacterium glutamicum ATCC13032 [28, 29] and riboflavin production in Bacillus subtilis RH33 [30]. To test the relationship between availability of NADPH and succinate production, plasmid pZY02E containing E. coli genes zwf-gnd and pZY02 containing deregulated genes zwf243-gnd361 from C. glutamicum under the control of inducible promoters Ptrc were constructed and transformed into a succinate producing strain ZTK (W1485, ΔptsG ldhA pflB) [31], creating strains ZTK (pZY02E) and ZTK (pZY02). Succinate yield by the resultant strain ZTK (pZY02E), at 1.12 mol/mol glucose, was 11 % higher than the control strain ZTK. However, the specific growth rate and specific glucose uptake rate decreased by 50 and 34 %, respectively. The intracellular metabolite analysis revealed that by-product acetate level was 15 % higher than ZTK (Table 1). As for strain ZTK (pZY02), the molar succinate yield reached the value of 1.18 mol/mol glucose, which was 17 % higher than that of strain ZTK and 5 % higher than that of strain ZTK (pZY02E), and the by-product acetate level was modestly reduced relative to the strain ZTK (pZY02E). The specific growth rate and specific glucose uptake rate decreased by 50 and 25 % compared to the corresponding measurements for ZTK, while the specific glucose uptake rate of ZTK (pZY02) improved by 14 % relative to the strain ZTK (pZY02E) (Table 1). Furthermore, an elevated generation of NADH was favorable for the utilization of deregulated zwf243-gnd361, the intracellular NADH/NAD+ ratio in ZTK (pZY02), at 0.46, was higher than the ratio 0.26 in ZTK and 0.36 in ZTK (pZY02E) (Fig. 2). It is evident from this experiment that increasing expression of zwf and gnd was positively correlated with the succinate yield, and recruiting deregulated zwf243 and gnd361 were more efficient to achieve a high succinate yield with significantly increased intracellular NADH availability.
Table 1.
Metabolic profiles of E.coli mutants cultivated in NBS medium supplemented with 10 g/L glucose under anaerobic condition
| Strains | Fermentation time (h) | CDW (g/L) | Glucose consumed (mM) | Succinate (mM) | Succinate yield (mol/mol) | Acetate (mM) | Pyruvate (mM) | Lactate (mM) | Specific growth rate during 0–24 h (g/g·h) | Specific glucose uptake rate during 0–24 h (g/g·h) |
|---|---|---|---|---|---|---|---|---|---|---|
| ZTK | 42 | 1.03 ± 0.07 | 54.26 ± 0.26 | 54.91 ± 0.29 | 1.01 ± 0.006 | 26.10 ± 1.49 | – | – | 0.042 ± 0.001 | 0.44 ± 0.01 |
| ZTK (pZY02E) | 84 | 0.75 ± 0.03 | 53.33 ± 0.45 | 59.56 ± 0.40 | 1.12 ± 0.004 | 30.1 ± 0.03 | – | – | 0.021 ± 0.002 | 0.29 ± 0.01 |
| ZTK (pZY02) | 72 | 0.79 ± 0.01 | 54.26 ± 0.26 | 64.08 ± 0.40 | 1.18 ± 0.006 | 28.18 ± 0.18 | – | – | 0.021 ± 0.001 | 0.33 ± 0.03 |
| WSA110 | 48 | 0.85 ± 0.01 | 53.89 ± 0.45 | 62.40 ± 0.38 | 1.16 ± 0.005 | 32.53 ± 0.62 | 1.50 ± 0.04 | – | 0.033 ± 0.0004 | 0.39 ± 0.03 |
| WSA134 | 48 | 0.83 ± 0.003 | 54.26 ± 0.26 | 65.80 ± 0.22 | 1.21 ± 0.002 | 31.48 ± 0.57 | – | – | 0.029 ± 0.001 | 0.37 ± 0.006 |
| WSA138 | 48 | 0.93 ± 0.008 | 54.44 ± 0.45 | 67.74 ± 0.36 | 1.24 ± 0.005 | 27.78 ± 0.56 | – | – | 0.035 ± 0.0008 | 0.43 ± 0.01 |
| WSA146 | 48 | 0.89 ± 0.05 | 53.33 ± 0.45 | 69.68 ± 0.26 | 1.31 ± 0.005 | 23.68 ± 0.28 | – | – | 0.032 ± 0.0007 | 0.40 ± 0.01 |
| WSA150 | 54 | 0.76 ± 0.008 | 51.48 ± 1.31 | 69.99 ± 1.55 | 1.36 ± 0.007 | 3.53 ± 0.04 | 1.44 ± 0.02 | 0.78 ± 0.06 | 0.031 ± 0.002 | 0.34 ± 0.03 |
| WSA152 | 48 | 0.75 ± 0.009 | 50.93 ± 0.26 | 66.75 ± 0.35 | 1.31 ± 0.001 | 1.74 ± 0.04 | 3.49 ± 0.11 | 0.49 ± 0.05 | 0.023 ± 0.002 | 0.38 ± 0.02 |
| WSA157 | 42 | 0.81 ± 0.014 | 51.67 ± 1.20 | 72.50 ± 1.59 | 1.40 ± 0.002 | 0.99 ± 0.03 | 1.71 ± 0.06 | 0.53 ± 0.09 | 0.032 ± 0.002 | 0.41 ± 0.01 |
| WSA159 | 48 | 0.69 ± 0.005 | 52.59 ± 0.26 | 80.56 ± 0.10 | 1.53 ± 0.006 | 1.36 ± 0.14 | 1.43 ± 0.04 | 0.85 ± 0.07 | 0.027 ± 0.002 | 0.39 ± 0.03 |
| WSA161 | 36 | 0.97 ± 0.02 | 50.56 ± 0.45 | 73.61 ± 0.70 | 1.46 ± 0.01 | 0.97 ± 0.17 | 0.96 ± 0.09 | 0.30 ± 0.03 | 0.051 ± 0.002 | 0.44 ± 0.03 |
| WSA163 | 60 | 0.72 ± 0.05 | 52.50 ± 0.26 | 80.72 ± 0.24 | 1.54 ± 0.01 | 1.34 ± 0.03 | 1.51 ± 0.02 | 0.96 ± 0.02 | 0.024 ± 0.005 | 0.28 ± 0.03 |
| WSA165 | 36 | 0.90 ± 0.03 | 52.59 ± 0.52 | 78.64 ± 0.79 | 1.50 ± 0.005 | 2.08 ± 0.05 | 2.00 ± 0.20 | – | 0.044 ± 0.002 | 0.44 ± 0.02 |
| WSA167 | 36 | 1.03 ± 0.02 | 53.33 ± 0.45 | 77.82 ± 0.55 | 1.46 ± 0.008 | 1.87 ± 0.06 | 2.66 ± 0.14 | – | 0.054 ± 0.002 | 0.54 ± 0.005 |
Bacteria were cultivated with 50 mL NBS medium in a 100 mL flask shaking at 220 rpm and 37 °C in anaerobic conditions with an initial OD600 of 0.6; “–” represents data were not measured in this work. Data are average values and standard deviations of triplicate experiments
Fig. 2.
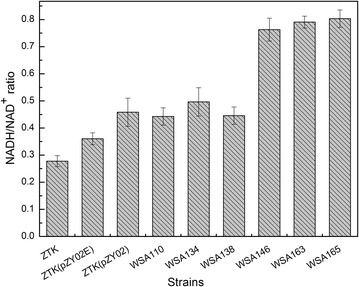
Changes in the levels of intracellular NADH/NAD+ ratio in different strains. Bacteria were cultivated with 50 mL NBS medium in a 100 mL flask shaking at 220 rpm and 37 °C in anaerobic conditions with an initial OD600 of 0.6. The intracellular NADH, NAD+ were extracted at exponential phase. Data are average values and standard deviations of triplicate experiments
However, plasmid overexpression has several disadvantages for engineering of genetically stable strains. Plasmid maintenance was a metabolic burden on the host cell, especially for high-copy number plasmids. In addition, only low-copy number plasmids had replication that was timed with the cell cycle, thus, it was difficult to maintain consistent copy number in all cells [13]. Taking these factors into account, we decided to implement all modulation of genes expression directly in chromosome in this study. We subsequently implemented zwf243 and gnd361 expression in chromosome of ZTK by replacing native lacZ encoding beta-D-galactosidase, resulting in strain WSA110. Zwf243 and gnd361 transcripts were up-regulated by 3.51 and 2.77-folds respectively under the inducible promoter Ptrc (Fig. 3). Succinate yield of this newly constructed strain increased to 1.16 mol/mol glucose, representing 15 % increase relative to the control strain ZTK. The intracellular NADH/NAD+ ratio in WSA110 was increased to 0.44 from 0.28 in ZTK (Fig. 2). While addition of IPTG (1 mM) dropped the specific growth rate and specific glucose uptake rate by 21 and 11 %, respectively. The main byproduct acetate was accumulated to 32.53 mM, which was higher than that of strain ZTK. In addition, a small amount of pyruvate had been detected (Table 1).
Fig. 3.
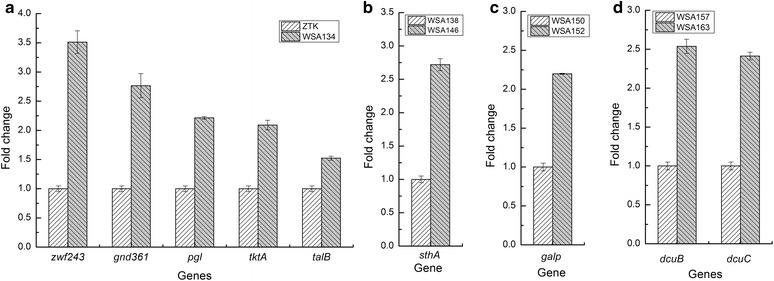
Results of relative transcriptional level. a The transcription level of the genes in PP pathway; b The transcription level of glucose uptake gene; c The transcription level of soluble transhydrogenase gene; d The transcription level of the genes in succinate export. Data are average values and standard deviations of triplicate experiments
In order to direct more carbon flow to PP pathway and further redirect flux to glycolic pathway, three critical enzymes of the PP pathway, 6-phosphogluconolactonase (encoded by pgl), transketolase (encoded by tktA) and transaldolase (encoded by talB) were overexpressed in WSA110 by replacing native promoter with constitutive promoter BJ23100 (http://www.parts.igem.org/Part:BBa_J23100). The transcript levels of genes pgl, tktA, talB were up-regulated by 2.2, 2.09, 1.52 folds respectively (Fig. 3). The intracellular NADH/NAD+ ratio in WSA134 was elevated to 0.50 (Fig. 2). As expected, succinate yield by the WSA134 was further increased, and the molar yield reached value of 1.21 mol/mol glucose, increasing by 20 % in comparison with strain ZTK and representing 71 % of the maximum stoichiometric yield. Meanwhile, the acetate level slightly reduced to 31.48 mM. The specific growth rate and specific glucose uptake rate of strain WSA134 were decreased by 12 and 5 % (Table 1).
Effect of overexpression of pepck from A. succinogenes and conversion of NADPH into NADH on succinate yield
The succinate yield was increased by 20 % thanks to modification of the PP pathway, while the acetate production showed a small increase and maintained in a higher level (Fig. 4). Diverting the flux towards succinate by ATP-forming phosphoenolpyruvate carboxykinase (PEPCK) should eliminate the need for the ATP producing acetate pathway, and thus could be used to further improve succinate formation [9]. Therefore, pepck (encoding PEPCK) from A. succinogenes was heterogenously expressed under the strong constitutive promoter BJ23100 by replacing native ppc. In response to this modification, cell concentration of resultant strain WSA138 increased by 12 %, and the specific growth rate and specific glucose uptake rate increased by 21 and 16 % in the anaerobic fermentation. While, the succinate yield increased slightly to 1.24 mol/mol glucose. Correspondingly, the titer of the main byproduct acetate reduced to 27.78 mM (Table 1). We speculated that excess NADPH produced by PP pathway was not transformed into NADH fast enough, thus insufficient NADH supply still limited succinate synthesis in WSA138. Overexpression of transhydrogenase SthA might be able to reinforce intracellular NADH level. In light of this speculation, the native promoter of sthA was replaced by the strong constitutive promoter BJ23100 in strain WSA138 to produce strain WSA146. The transcript levels of gene sthA was up-regulated by 2.72-folds (Fig. 3). As expected, the intracellular NADH/NAD+ ratio was distinctly increased to 0.76 from 0.45 in WSA138 (Fig. 2). The succinate yield increased to 1.31 mol/mol glucose, which represents 76 % of the maximum stoichiometric yield. However, the formation of the main byproduct acetate decreased to 23.68 mM (Table 1).
Fig. 4.
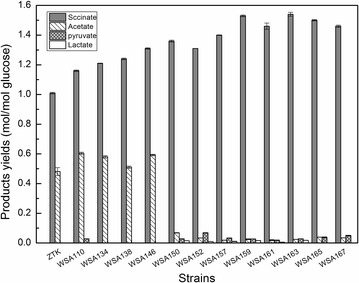
Comparison of the products yields of different mutants. Bacteria were cultivated with 50 mL NBS medium in a 100 mL flask shaking at 220 rpm and 37 °C in anaerobic conditions with an initial OD600 of 0.6. Data are average values and standard deviations of triplicate experiments
Deletion of the ackA-pta gene leads to increased succinate yield at expense of reduced glucose consumption
Compared to control ZTK, a 30 % increase in terms of succinate yield was achieved by activating PP pathway, overexpressing sthA and employing PEPCK as the primary CO2-fixation enzyme (Table 2), whereas acetate was always found to be the major byproduct. The presence of acetate at high concentrations is undesirable in the fermentation broth since it interferes with the target succinate biosynthesis and increases the cost of succinate purification [32]. Thus, it is essential to remove acetate formation. Phosphotransacetylase (PTA) and acetate kinase (ACKA), encoded by the pta and ackA genes respectively, are the major players in acetate synthesis under oxygen deprivation [15]. Deletion of ackA-pta resulted in strain WSA150, of which acetate production decreased about sixfold and succinate yield increased to 1.36 mol/mol glucose with trace amount of lactate and pyruvate being produced (Table 1). Unfortunately, this yield was obtained at the expense of reduction in specific glucose uptake rate and biomass concentration, each representing 15 and 15 % decrease compared to strain WSA146 (Table 1).
Table 2.
Comparison of molar yields of succinate in anaerobic fermentation
| Strains | Yield of succinate (mol/mol) | Increased yield compared with ZTK (%) | Yield compared with the theoretical maximum yield (%) |
|---|---|---|---|
| ZTK | 1.01 ± 0.006 | 0 | 59 |
| WSA134 | 1.21 ± 0.002 | 20 | 71 |
| WSA146 | 1.31 ± 0.005 | 30 | 76 |
| WSA157 | 1.40 ± 0.002 | 39 | 82 |
| WSA159 | 1.53 ± 0.006 | 51 | 89 |
| WSA163 | 1.54 ± 0.010 | 52 | 90 |
| WSA165 | 1.50 ± 0.005 | 49 | 88 |
Bacteria were cultivated with 50 mL NBS medium in a 100 mL flask shaking at 220 rpm and 37 °C in anaerobic conditions with an initial OD600 of 0.6. Data are average values and standard deviations of triplicate experiments
Overexpressing galp gene increases glucose consumption with compromising succinate yield
Disruption of ackA-pta greatly reduced acetate synthesis, but the mutant strain cultured in minimal medium exhibited low growth and glucose consumption rates. Galactose permease (encoded by galp) of E. coli was engineered to recover glucose uptake capacity and increase succinate productivity as previously reported [13, 33]. In this work, galp was carried out in chromosome with substitution of strong constitutive promoter BJ23100 for native promoter to improve anaerobic glucose transport velocity. The transcript level of gene galp was up-regulated by 2.2 folds (Fig. 3). The resultant strain WSA152 was characterized by reduced fermentation time, and its specific glucose uptake rate increased by 12 %. The specific growth rate of WSA152 had a modest reduction, demonstrating that WSA152 had an advantage over WSA150 in terms of individual cell glucose consumed rate. Nevertheless, the succinate yield decreased to 1.31 mol/mol glucose and byproduct pyruvate increased about 1.4-fold to 3.49 mM (Table 1). Moreover, the fermentation broth accumulated little amounts of acetate and lactate. The reasons for this phenomenon may be attributed to that the intracellular glucose input flux is not efficiently channeled into succinate, and in turn leads to increased cell overflow metabolism after galp overexpression.
Overexpressing pyc from C. glutamicum enhances glucose consumption and succinate yield
The reaction catalyzed by pyruvate carboxylase (PYC) functions as anaplerotic pathways for replenishing oxaloacetate (OAA) under anaerobic conditions [10]. Recruiting heterologous PYC is beneficial to suppress pyruvate formation and further improve succinate yield [34]. Herein, pyc encoding pyruvate carboxylase from C. glutamicum was introduced into WSA152 by replacing native lldD encoding L-lactate dehydrogenase. As expected, the succinate yield elevated to 1.40 mol/mol glucose (amounting to 82 % of the maximum stoichiometric yield) with reducing byproduct pyruvate by half. In addition, trace amount of lactate and acetate were detected in the fermentation broth. Notably, the specific growth rate increased by 39 % and specific glucose uptake rate increased by 8 % in response to PYC recruit (Table 1). The higher succinate yield of WSA157 resulted from the combined effects of increased glucose consumption and suppressed pyruvate production due to an alternative route to OAA pool provided by pyc introduction.
Overexpressing dcuB and dcuC genes to improve succinate yield close to the stoichiometric maximum yield
Escherichia coli contains four Dcu carriers (encoded by dcuA, dcuB, dcuC, and dcuD genes) for the transportation of C4-dicarboxylates (succinate, fumarate, and malate) under anaerobic conditions [35, 36]. The uptake, antiport, and export of C4-dicarboxylates are mediated by the Dcu transporters [37]. As decreasing intracellular succinate is an effective strategy for increasing succinate production, we additionally targeted to activate anaerobic C4-dicarboxylates efflux transportation to maximize succinate yield. Therefore, upstream regulated region of the dcuB and dcuC operons in WSA157 were replaced with strong constitutive promoter BJ23100, respectively, resulting in strains WSA159 and WSA161. Succinate production by the strain WSA159 increased to 80.56 mM, and the molar yield reached the value of 1.53 mol/mol glucose, which is 89 % of the strain-specific stoichiometric maximum. Correspondingly, concomitant decrease in cell growth and glucose consumption was observed for WSA159. Its specific growth rate decreased by 16 % and specific glucose uptake rate decreased by 5 % compared to WSA157 (Table 1). For strain WSA161, this modification increased succinate yield to 1.46 mol/mol glucose, amounting to 85 % of stoichiometric maximum yield. It is worth noting that dcuC overexpression increased the specific growth rate by 59 % and specific glucose uptake rate by 7 % over the corresponding measurements for WSA157 (Table 1). These results demonstrated that activation of Dcu transportations effectively accelerated the anaerobic succinate transport out of E. coli.
In order to investigate whether the activation of both DcuB and DcuC had a synergistic effect in improving succinate yield, genes dcuB and dcuC were simultaneously overexpressed in strain WSA157, creating strain WSA163. The transcript levels of genes dcuB and dcuC of were up-regulated by 2.54 and 2.41-folds respectively (Fig. 3). The strain WSA163 synthesized succinate with a molar yield of 1.54 mol/mol glucose (90 % of corresponding stoichiometric maximum), which was similar to that of strain WSA159. Nevertheless, the specific growth rate of WSA163 decreased by 25 % and specific glucose uptake rate decreased by 32 % compared to the parent strain WSA157 (Table 1). The intracellular NADH/NAD+ ratio in WSA163 was modestly raised (Fig. 2).
Deletion of the repressor protein binding site leads to removed IPTG induction
In the present work, all genes overexpression was modulated by constitutive promoter BJ23100 with the exception of genes zwf243 and gnd361, which were modulated by the IPTG-induced trc promoter. It is not feasible for large-scale production of bulk chemicals and biomass growth with the IPTG induced expression. The repressor protein binding site (AATTGTGAGCGGATAACAATT) was deleted in the two high succinate yield strains WSA159 and WSA163, resulting in strains WSA165 and WSA167, respectively. It was found that deletion the repressor protein binding site dramatically improved final cell concentration. The specific growth rate and specific glucose uptake rate of WSA165 increased by 63 and 13 %, while the succinate yield decreased slightly and maintained at 1.50 mol/mol glucose, which represents 88 % of the maximum stoichiometric yield. The intracellular NADH/NAD+ ratio of strain WSA165 still maintained at a high level (Fig. 2). Moreover, this deletion increased the specific growth rate of WSA167 more than one fold and specific glucose uptake rate by 93 % over the corresponding measurements under oxygen deprivation, while its succinate yield decreased to 1.46 mol/mol glucose. In addition, a small amount of pyruvate and acetate had been detected in the media of both strains (Table 1).
Discussion
Succinate synthesis through the reductive branch of the TCA cycle with the CO2 fixation is an attractive option under anaerobic conditions [4, 24]. However, the maximum theoretical yield of succinate through the glycolysis and reductive TCA cycle is 1 mol/mol glucose resulting from the NADH limitation [14, 24, 38]. It is well known that the excess NADH would upset the flux distribution among the end products: succinate, acetate and ethanol [9]. There is no excess NADH to be oxidized by producing lactate or ethanol in the ZTK strain (W1485, ΔptsG ldhA pflB), while NADH supply is still limited for higher yield succinate production. In this work, PP pathway was verified for the first time to provide more NADH for succinate production compared with the traditional glycolysis pathway. On the premise that NADPH produced by PP pathway can be immediately converted into NADH through transhydrogenase SthA, the stoichiometric maximum succinate yield of 1.714 mol/mol glucose can be obtained if the carbon flux ratio between PP pathway and glycolysis is 6:1. Based on this theoretical guidance, we deregulated the PP pathway and glycolytic pathway flux ratio and further strengthened the conversion of NADPH into NADH. We systematically adjusted the expression of five critical enzymes of PP pathway for succinate production. The deregulated genes zwf243 and gnd361 involved in NADPH generation from C. glutamicum were firstly introduced into E. coli for succinate production for the first time. We compared the effects of overexpression of native zwf-gnd and deregulated zwf243-gnd361 on succinate production and NADH/NAD+ ratio. It was found that co-expression of the mutated dehydrogenases, which removed allosteric inhibition by intracellular metabolites, was more efficient to achieve a high succinate yield with significantly increased intracellular NADH availability. An improvement by 15 % of the succinate yield was obtained by overexpressing of the deregulated zwf243 and gnd361 directly in chromosome of ZTK. In this study, increasing carbon flux towards PP pathway through expression of deregulated zwf243 and gnd361 was investigated in strain ZTK (W1485, ΔptsG ldhA pflB), and based on this genetic background, high-yield succinate production in strain ZTK was restricted by multiple limiting factors, including NADH availability, carbon flux towards succinate, succinate export, and etc. It is reasonable that only a minor contribution to the succinate yield improvement (increased by 15 %) by introduction of deregulated zwf243 and gnd361 into strain ZTK. An extended application of mutant zwf243 and gnd361 is still recommended for the production of products directly stemming from the PP pathway as well as other NADH-demanding compounds in E. coli. Three critical genes of PP pathway were then overexpressed to further divert more carbon flux to the PP pathway. The engineered strain WSA134 by disturbing PP pathway achieved succinate yield of 1.21 mol/mol glucose, increased 20 % compared to ZTK, amounting to 71 % of theoretical maximum yield (Table 2). These results indicated that more carbon flux went through the PP pathway, thus leading to production of more reducing equivalent in the form of NADPH, which was then converted into NADH through transhydrogenase SthA for succinate synthesis.
In addition, for the production of succinate, one of the key factors is the supply of CO2 [4]. If all carbon flux goes through glycolysis/reductive TCA cycle, the maximum theoretical yield of succinate resulting from the NADH limitation is based on the reaction: C6H12O6 + CO2 → C4H6O4 + H2O, in this case one CO2 is required for the synthesis of one succinate. By contrast, if 1/7 carbon flux goes through glycolysis/reductive TCA cycle and 6/7 carbon flux goes through the combined PP pathway and glycolysis/reductive TCA route to meet the NADH requirement, the maximum theoretical yield of succinate is based on the stoichiometry of the following redox reaction: C6H12O6 + 0.857 CO2 → 1.714 C4H6O4 + 0.857 H2O, so in this case only 0.5 (0.857/1.714) CO2 is required for the synthesis of one succinate. Apparently, less exogenous CO2 is required for succinate synthesis using PP pathway.
Carbon fluxes at the PEP node serve to limit the amount of succinate produced for redox balance during the anaerobic glucose fermentation. Besides, these fluxes must provide sufficient energy (ATP) for growth, maintenance, and precursors for succinate biosynthesis [11]. In this work, we carried out a theoretical metabolic flux balance analysis (FBA) [39] of succinate production by E. coli under anaerobic conditions with/without ppc replacement by pepck. Flux distributions computed by FBA (data not shown) showed that sole production of succinate through the reductive TCA cycle does not lead to ATP generation without ppc replacement by pepck, ATP production is mainly associated with acetate formation, but the presence of acetate at high concentrations is undesirable in the fermentation broth since it interferes with the target succinate biosynthesis and increases the cost of succinate purification [32]. When native ppc is replaced by A. succinogenes pepck, the conversion of glucose to succinate will lead to net production of ATP. With the presence of PEPCK, the ATP production associated with succinate production corresponding to the maximum theoretical yield of succinate is based on the reaction: C6H12O6 + 0.857 CO2 → 1.714 C4H6O4 + 1.714 ATP + 0.857 H2O, the conversion of 1 mol glucose to succinate will produce 1.714 mol ATP. Diverting the flux towards succinate by ATP-forming PEPCK should increase carbon flow into the four-carbon intermediate OAA for succinate production, increase the net production of ATP for growth and maintenance and eliminate the need for the ATP producing acetate pathway [11]. In our existing work, pepck overexpression indeed increased the specific growth rate, specific glucose uptake rate and biomass by 21, 16 and 12 %, respectively, and acetate level decreased from 31.48 to 27.78 mM (Table 1). While this modification did not result in significant increase in succinate yield, this might be because excess NADPH was not converted into NADH fast enough. The succinate yield was further increased through sthA overexpression. Apparently, transhydrogenase SthA plays an important role in terms of NADH supply. It should be a potential strategy to regulate the NADH generation for succinate synthesis through combining action of the PP pathway and transhydrogenase SthA.
Deletion ackA-pta promoted succinate yield but decreased the cells’ overall fitness. Heterologous expression of PYC from C. glutamicum led to higher succinate yield, and restored the ability of the engineered strain to ferment glucose. 90 % of stoichiometric maximum succinate yield was achieved after modulating succinate efflux transportation. The deletions of repressor protein binding sequences led to the constitutive expression of the PP pathway genes zwf243-gnd361, and the engineered strains were observed to increase cellular fitness during anaerobic fermentation. Future engineering efforts could focus on modulating the delicate split of the carbon flux between PP pathway and glycolytic pathway, thus leading to achieve succinate yield close to the target value of 1.714 mol/mol glucose.
Conclusions
For the first time, pentose phosphate pathway was rationally designed to improve NADH supply for efficient succinate yield in E. coli under anaerobic conditions. Through further combing diverting flux towards succinate and engineering its export, the succinate yield reached the value of 1.54 mol/mol glucose, representing 90 % of stoichiometric maximum. The systems metabolic engineering strategy described in this work may also be useful for the synthesis of other related NADH-demanding products.
Methods
Bacterial strains, primers, and media
Bacterial strains and plasmids used in this study were listed in Table 3. All the primers used in this study were listed in Additional file 1: Table S1. E. coli ZTK (W1485; ΔptsG ΔldhA ΔpflB) [31] was used as the parent strain for construction of all mutants described in this study. E. coli DH5α was used as the host for plasmid construction. E. coli cells were cultivated at 37 or 30 °C in a rotatory shaker at 220 rpm in Luria–Bertani (LB). Ampicillin (100 µg/mL), tetracycline (100 µg/mL), spectinomycin (100 µg/mL) and chloramphenicol (16 µg/mL) were added as needed.
Table 3.
Strains and plasmids used in this study
| Strains and plasmids | Relevant characteristics | Sources or references |
|---|---|---|
| Strains | ||
| ZTK | W1485, ΔptsG ΔldhA ΔpflB; kanr | Lab stock |
| WSA110 | ZTK, ΔlacZ::Ptrc-zwf243-gnd361 from C. glutamicum; kanr | This study |
| WSA134 | WSA110, PBJ23100-talB; PBJ23100-tktA; PBJ23100-pgl; kanr | This study |
| WSA138 | WSA134, PBJ23100-pepck; Δppc::PBJ23100-pepck from A. succinogenes; kanr | This study |
| WSA146 | WSA138, PBJ23100-sthA; kanr | This study |
| WSA150 | WSA146, ΔackA-pta; kanr | This study |
| WSA152 | WSA150, PBJ23100-galp; kanr | This study |
| WSA157 | WSA152, ΔlldD::PBJ23100-pyc from C. glutamicum; kanr | This study |
| WSA159 | WSA157, PBJ23100-dcuB; kanr | This study |
| WSA161 | WSA157, PBJ23100-dcuC; kanr | This study |
| WSA163 | WSA157, PBJ23100-dcuB; PBJ23100-dcuC; kanr | This study |
| WSA165 | WSA159, Δzwf243-gnd361-protein bind; kanr | This study |
| WSA167 | WSA163, Δzwf243-gnd361-protein bind; kanr | This study |
| DH5α | Cloning host | Lab stock |
| Plasmids | ||
| pZY02E | p15 ori; Ptrc-zwf-gnd of E.coli; Cmr | This study |
| pZY02 | p15 ori; Ptrc-zwf243-gnd361 of C. glutamicum; Cmr | Lab stock |
| pCU18 | pBR322 ori; Ampr | Lab stock |
| pCU18-zwf243-gnd361 | Ptrc-zwf243-gnd361 from pZY02, cat gene from pEL04, upstream and downstream fragments of lacz gene were cloned into pCU18; Ampr, Cmr | This study |
| pCU18-pyc | PBJ23100- pyc from C. glutamicum was cloned into pCU18; Ampr | This study |
| pCU18-ppc | Upstream and downstream fragments of ppc gene were cloned into pCU18; Ampr | This study |
| pCU18-cat-sacB | cat-sacB genes from pEL04 were cloned into pCU18-ppc; Ampr, Cmr | This study |
| pCU18-pepck | PBJ23100- pepck from A. succinogenes was cloned into pCU18-ppc; Ampr | This study |
| pEL04 | cat-sacB cassette; Cmr | [41] |
| PTKSS | p15A replication, Tetr, I-SceI restriction sites; Cmr, Tetr | [49] |
| PTKRED | pSC10 replication, temperature sensitive replication origin, ParaBAD-driven I-SceI gene, red recombinase expression plasmid, lac-inducible expression; Spcr | [49] |
kan kanamycin; Amp ampicillin; Cm chloramphenicol; Tet tetracycline; Spc spectinomycin; r resistance
Plasmid construction
To construct pZY02E, zwf, gnd genes from E. coli with the synthesized ribosome binding sites (RBSs) were amplified by polymerase chain reaction (PCR) using the primers pzw-F/pzw-R, pgd-F/pgd-R respectively. Primers pzy-F/pzy-R were used to amplify the backbone of the plasmid pZY02. The zwf-gnd genes of E. coli were cloned into pZY02 by replacing zwf243-gnd361 genes to yield pZY02E. The method used for the construction of pZY02E was based on the utilization of circular polymerase extension cloning (CPEC) [40].
To construct pCU18-zwf243-gnd361, upstream and downstream fragments of lacz gene were amplified from E. coli genome using primers lacz-up-F/lacz-up-R and lacz-down-F/lacz-down-R, cat gene and zwf243-gnd361 genes were amplified from pEL04 [41] and pZY02 (containing zwf243 and gnd361 mutation under trc promoter) using primers cm-F/cm-R and zw-F/zw-R, respectively. The upstream and cat gene fragments were fused and amplified by fusion PCR with primers lacz-up-F/cm-R, three fragments were then digested with SmaI-SacI, HindIII-PstI, SalI-SmaI respectively and ligated into pCU18 digested with the same enzymes to create pCU18-zwf243-gnd361.
For the construction of pCU18-pyc, the fragments of pyc gene with promoter and ribosomal binding site of E. coli was amplified from C. glutamicum genome using primers pyc-F/pyc-R and digested with SphI- SacI. The digested fragment was inserted into pCU18 with the same enzymes to yield pCU18-pyc.
For pCU18-cat-sacB construction, the ppc and cat-sacB fragments were amplified from E. coli genome and pEL04 using primers ppc-up-F/ppc-down-R and cat-F/sacB-R respectively. The ppc fragment was digested with HindIII- SmaI and ligated into pCU18 digested with the same enzymes to yield pCU18-ppc. The obtained plasmid was digested with MIuI-SphI and ligated with cat-sacB fragment digested with the same restriction enzymes to give plasmid pCU18-cat-sacB.
For pCU18-pepck construction, pepck fragments was amplified from A. succinogenes genome using primers pck-F/pck-R. The PCR product was digested with MIuI-SphI and inserted into the same sites of pCU18-ppc to yield pCU18-pepck.
Strain construction
Chromosomal modifications were performed using the method of λ-red recombination with the corresponding recombinant fragments. The final fragments were transformed into the competent cells with expression of the λ-red recombination enzymes, and the procedure in detail was achieved as described previously [42, 43]. The method used for the integration of pyc in the chromosome of E. coli was based on the utilization of CRISPR–Cas9 meditated genome editing system [44, 45].
Anaerobic fermentation
The composition of the NBS medium used for succinate production was achieved as described previously [46]. All fermentations were carried out at 37 °C in a rotatory shaker at 220 rpm. Single bacterial colonies were grown in a 15 ml glass tube containing 5 ml of LB medium for 8 h. The culture was then inoculated into 500 mL flask containing 90 mL LB medium with an initial OD600 about 0.05 and then cultured for 5 h. Cells were harvested by centrifugation for 5 min at 6000 rpm and 4 °C, washed once with 50 mL NBS and used as an inoculum for the fermentation. The fermentation was carried out in triplets, in sealed 100-mL bottles containing 50 mL of NBS medium supplement with 10 g/L glucose, 100 mM NaHCO3. 1 mM of isopropyl-β-D-thiogalactopyranoside (IPTG) was added at the beginning of anaerobic culture to explore the appropriate expression quantity of mutant genes zwf243 and gnd361 for succinate production. At the start of the dual-phase fermentation, the cells were grown in the existed small amount of air which keeps a short time of microaerobic condition and was consumed rapidly.
Analytical techniques
Cell growth was monitored by measuring the OD600 with an ultraviolet spectrophotometer (Beijing Puxi Universal Co. Ltd). The biomass concentration was calculated from OD600 values using an experimentally determined correlation with 1 OD600 unit being equal to 0.38 g/L cell dry weight (CDW) [42]. Glucose in the fermentation broth was determined utilizing a SBA sensor machine (Institute of Microbiology, Shangdong, China). To determine succinate, pyruvate, acetate and lactate concentrations, culture samples were centrifuged at 12,000g for 5 min and the aqueous supernatant used for HPLC analysis on an Agilent 1100 Series HPLC system equipped with a cation exchange column. (Aminex HPX87-H, Bio-Rad, Hercules, CA, USA), a UV absorbance detector (Agilent Technologies, G1315D) and a refractive index (RI) detector (Agilent Technologies, HP1047A). A mobile phase of 5 mM H2SO4 solution at a 0.4 mL/min flow rate was used. The column was operated at 65 °C. The intracellular NADH and NAD+ levels were determined by NAD/NADH Quantitation Kit (SIGMA) [47].
Quantitative real-time reverse transcription (RT)-PCR analysis
Escherichia coli ZTK and its derivatives were harvested when grown to the exponential phase under anaerobic condition. Total RNA was extracted with RNAprep pure Kit (Tiangen, Beijing, China). 500 ng of total RNA was transcribed into cDNA using Quant Reverse Transcriptase with random primers (Tiangen, Beijing, China). Samples were then analyzed using a Light Cycler 480 II (Roche, Basel, Switzerland) with Real Master Mix (SYBR Green). The 16S rRNA gene was selected as reference for normalization and three biological replicates were performed. The obtained data were analyzed by using the 2−ΔΔCt method previously described [48].
Authors’ contributions
JM and BW performed the experiments under the guidance of ZW and XZ; JM, BW and ZW analyzed the experimental data and drafted the manuscript. TC and DL made substantial contributions to conception, interpretation of data and revised the manuscript. ZW, JM and BW developed the idea for the study and designed the research. All authors read and approved the final manuscript.
Acknowledgements
None.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datas supporting the conclusions of this article are all available in the manuscript and supplementary.
Funding
This work was supported by National Program on Key Basic Research Project (2012CB725203), National Natural Science Foundation of China (NSFC-21576200 and NSFC-21390201), National High-tech R&D Program of China (2012AA02A702) and Natural Science Foundation of Tianjin (No. 15JCQNJC06000).
Abbreviations
- PP
pentose phosphate
- TCA
reductive tricarboxylic acid
- GldA
glycerol dehydrogenase
- FBP
fructose 1,6-bisphosphate
- Gra3P
D-glyceraldehyde 3-phosphate
- PEPCK
phosphoenolpyruvate carboxykinase
- PTA
phosphotransacetylase
- ACKA
acetate kinase
- PYC
pyruvate carboxylase
- OAA
oxaloacetate
- IPTG
isopropyl-β-D-thiogalactopyranoside
- CDW
cell dry weight
- RI
refractive index
Additional file
10.1186/s12934-016-0536-1 Primer sequence used in this study.
Footnotes
Jiao Meng and Baiyun Wang contributed equally to this work
Contributor Information
Jiao Meng, Email: jiaomeng@tju.edu.cn.
Baiyun Wang, Email: baiyun@tju.edu.cn.
Dingyu Liu, Email: dingyuliu@tju.edu.cn.
Tao Chen, Email: chentao@tju.edu.cn.
Zhiwen Wang, Phone: +86-22-85356617, Email: zww@tju.edu.cn.
Xueming Zhao, Email: xmzhao@tju.edu.cn.
References
- 1.McKinlay JB, Vieille C, Zeikus JG. Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol. 2007;76:727–740. doi: 10.1007/s00253-007-1057-y. [DOI] [PubMed] [Google Scholar]
- 2.Zeikus JG. Chemical and fuel production by anaerobic bacteria. Annu Rev Microbiol. 1980;34:423–464. doi: 10.1146/annurev.mi.34.100180.002231. [DOI] [PubMed] [Google Scholar]
- 3.Samuelov NS, Lamed R, Lowe S, Zeikus JG. Influence of CO(2)-HCO(3) levels and pH on growth, succinate production, and enzyme activities of Anaerobiospirillum succiniciproducens. Appl Environ Microbiol. 1991;57:3013–3019. doi: 10.1128/aem.57.10.3013-3019.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou W, Zhu LW, Li HM, Tang YJ. Significance of CO2 donor on the production of succinic acid by Actinobacillus succinogenes ATCC 55618. Microb Cell Fact. 2011;10:87. doi: 10.1186/1475-2859-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SJ, Song H, Lee SY. Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Appl Environ Microbiol. 2006;72:1939–1948. doi: 10.1128/AEM.72.3.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark DP. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989;5:223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- 7.Jantama K, Zhang X, Moore JC, Shanmugam KT, Svoronos SA, Ingram LO. Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng. 2008;101:881–893. doi: 10.1002/bit.22005. [DOI] [PubMed] [Google Scholar]
- 8.Stols L, Donnelly MI. Production of succinic acid through overexpression of NAD(+)-dependent malic enzyme in an Escherichia coli mutant. Appl Environ Microbiol. 1997;63:2695–2701. doi: 10.1128/aem.63.7.2695-2701.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh A, Cher Soh K, Hatzimanikatis V, Gill RT. Manipulating redox and ATP balancing for improved production of succinate in E. coli. Metab Eng. 2011;13:76–81. doi: 10.1016/j.ymben.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Vemuri GN, Eiteman MA, Altman E. Effects of growth mode and pyruvate carboxylase on succinic acid production by metabolically engineered strains of Escherichia coli. Appl Environ Microbiol. 2002;68:1715–1727. doi: 10.1128/AEM.68.4.1715-1727.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Jantama K, Moore JC, Jarboe LR, Shanmugam KT, Ingram LO. Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proc Natl Acad Sci USA. 2009;106:20180–20185. doi: 10.1073/pnas.0905396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan Z, Zhu X, Chen J, Li Q, Zhang X. Activating phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in combination for improvement of succinate production. Appl Environ Microbiol. 2013;79:4838–4844. doi: 10.1128/AEM.00826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, Tang J, Liu Y, Zhu X, Zhang T, Zhang X. Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl Microbiol Biotechnol. 2012;93:2455–2462. doi: 10.1007/s00253-011-3752-y. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez AM, Bennett GN, San KY. Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab Eng. 2005;7:229–239. doi: 10.1016/j.ymben.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Zhu LW, Li XH, Zhang L, Li HM, Liu JH, Yuan ZP, Chen T, Tang YJ. Activation of glyoxylate pathway without the activation of its related gene in succinate-producing engineered Escherichia coli. Metab Eng. 2013;20:9–19. doi: 10.1016/j.ymben.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Jantama K, Haupt MJ, Svoronos SA, Zhang X, Moore JC, Shanmugam KT, Ingram LO. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol Bioeng. 2008;99:1140–1153. doi: 10.1002/bit.21694. [DOI] [PubMed] [Google Scholar]
- 17.Zhu X, Tan Z, Xu H, Chen J, Tang J, Zhang X. Metabolic evolution of two reducing equivalent-conserving pathways for high-yield succinate production in Escherichia coli. Metab Eng. 2014;24:87–96. doi: 10.1016/j.ymben.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Vemuri GN, Eiteman MA, Altman E. Succinate production in dual-phase Escherichia coli fermentations depends on the time of transition from aerobic to anaerobic conditions. J Ind Microbiol Biotechnol. 2002;28:325–332. doi: 10.1038/sj.jim.7000250. [DOI] [PubMed] [Google Scholar]
- 19.Liang L, Liu R, Wang G, Gou D, Ma J, Chen K, Jiang M, Wei P, Ouyang P. Regulation of NAD(H) pool and NADH/NAD(+) ratio by overexpression of nicotinic acid phosphoribosyltransferase for succinic acid production in Escherichia coli NZN111. Enzyme Microb Technol. 2012;51:286–293. doi: 10.1016/j.enzmictec.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Jantama K, Shanmugam KT, Ingram LO. Reengineering Escherichia coli for succinate production in mineral salts medium. Appl Environ Microbiol. 2009;75:7807–7813. doi: 10.1128/AEM.01758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blankschien MD, Clomburg JM, Gonzalez R. Metabolic engineering of Escherichia coli for the production of succinate from glycerol. Metab Eng. 2010;12:409–419. doi: 10.1016/j.ymben.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Shanmugam KT, Ingram LO. Fermentation of glycerol to succinate by metabolically engineered strains of Escherichia coli. Appl Environ Microbiol. 2010;76:2397–2401. doi: 10.1128/AEM.02902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balzer GJ, Thakker C, Bennett GN, San KY. Metabolic engineering of Escherichia coli to minimize byproduct formate and improving succinate productivity through increasing NADH availability by heterologous expression of NAD-dependent formate dehydrogenase. Metab Eng. 2013;20C:1–8. doi: 10.1016/j.ymben.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Skorokhodova AY, Morzhakova AA, Gulevich AY, Debabov VG. Manipulating pyruvate to acetyl-CoA conversion in Escherichia coli for anaerobic succinate biosynthesis from glucose with the yield close to the stoichiometric maximum. J Biotechnol. 2015;214:33–42. doi: 10.1016/j.jbiotec.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Park SH, Kim HU, Kim TY, Park JS, Kim SS, Lee SY. Metabolic engineering of Corynebacterium glutamicum for l-arginine production. Nat Commun. 2014;5:4618. doi: 10.1038/ncomms5618. [DOI] [PubMed] [Google Scholar]
- 26.Lee WH, Kim MD, Jin YS, Seo JH. Engineering of NADPH regenerators in Escherichia coli for enhanced biotransformation. Appl Microbiol Biotechnol. 2013;97:2761–2772. doi: 10.1007/s00253-013-4750-z. [DOI] [PubMed] [Google Scholar]
- 27.Cao Z, Song P, Xu Q, Su R, Zhu G. Overexpression and biochemical characterization of soluble pyridine nucleotide transhydrogenase from Escherichia coli. FEMS Microbiol Lett. 2011;320:9–14. doi: 10.1111/j.1574-6968.2011.02287.x. [DOI] [PubMed] [Google Scholar]
- 28.Ohnishi J, Katahira R, Mitsuhashi S, Kakita S, Ikeda M. A novel gnd mutation leading to increased l-lysine production in Corynebacterium glutamicum. FEMS Microbiol Lett. 2005;242:265–274. doi: 10.1016/j.femsle.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Becker J, Klopprogge C, Herold A, Zelder O, Bolten CJ, Wittmann C. Metabolic flux engineering of l-lysine production in Corynebacterium glutamicum–over expression and modification of G6P dehydrogenase. J Biotechnol. 2007;132:99–109. doi: 10.1016/j.jbiotec.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Chen T, Ma X, Shen Z, Zhao X. Enhancement of riboflavin production with Bacillus subtilis by expression and site-directed mutagenesis of zwf and gnd gene from Corynebacterium glutamicum. Bioresour Technol. 2011;102:3934–3940. doi: 10.1016/j.biortech.2010.11.120. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Z, Chen T, Zhao M, Wang Z, Zhao X. Engineering Escherichia coli for succinate production from hemicellulose via consolidated bioprocessing. Microb Cell Fact. 2012;11:37. doi: 10.1186/1475-2859-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sánchez AM, Bennett GN, San KY. Efficient succinic acid production from glucose through overexpression of pyruvate carboxylase in an Escherichia coli alcohol dehydrogenase and lactate dehydrogenase mutant. Biotechnol Prog. 2005;21:358–365. doi: 10.1021/bp049676e. [DOI] [PubMed] [Google Scholar]
- 33.Tang J, Zhu X, Lu J, Liu P, Xu H, Tan Z, Zhang X. Recruiting alternative glucose utilization pathways for improving succinate production. Appl Microbiol Biotechnol. 2013;97:2513–2520. doi: 10.1007/s00253-012-4344-1. [DOI] [PubMed] [Google Scholar]
- 34.Lin H, San KY, Bennett GN. Effect of Sorghum vulgare phosphoenolpyruvate carboxylase and Lactococcus lactis pyruvate carboxylase coexpression on succinate production in mutant strains of Escherichia coli. Appl Microbiol Biotechnol. 2005;67:515–523. doi: 10.1007/s00253-004-1789-x. [DOI] [PubMed] [Google Scholar]
- 35.Zientz E, Janausch IG, Six S, Unden G. Functioning of DcuC as the C4-dicarboxylate carrier during glucose fermentation by Escherichia coli. J Bacteriol. 1999;181:3716–3720. doi: 10.1128/jb.181.12.3716-3720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janausch I, Kim O, Unden G. DctA- and Dcu-independent transport of succinate in Escherichia coli: contribution of diffusion and of alternative carriers. Arch Microbiol. 2001;176:224–230. doi: 10.1007/s002030100317. [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Zhu X, Tan Z, Xu H, Tang J, Xiao D, Zhang X. Activating C4-dicarboxylate transporters DcuB and DcuC for improving succinate production. Appl Microbiol Biotechnol. 2014;98:2197–2205. doi: 10.1007/s00253-013-5387-7. [DOI] [PubMed] [Google Scholar]
- 38.Singh A, Lynch MD, Gill RT. Genes restoring redox balance in fermentation-deficient E. coli NZN111. Metab Eng. 2009;11:347–354. doi: 10.1016/j.ymben.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quan J, Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Lin Z, Liu Q, Li Y, Wang Z, Ma H, Chen T, Zhao X. Engineering of serine-deamination pathway, entner-doudoroff pathway and pyruvate dehydrogenase complex to improve poly (3-hydroxybutyrate) production in Escherichia coli. Microb Cell Fact. 2014;13:1–11. doi: 10.1186/1475-2859-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pines G, Freed EF, Winkler JD, Gill RT. Bacterial recombineering: genome engineering via phage-based homologous recombination. ACS Synth Biol. 2015;4:1176–1185. doi: 10.1021/acssynbio.5b00009. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Lin Z, Huang C, Zhang Y, Wang Z, Tang YJ, Chen T, Zhao X. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Pines G, Pines A, Garst AD, Zeitoun RI, Lynch SA, Gill RT. Codon compression algorithms for saturation mutagenesis. ACS Synth Biol. 2015;4:604–614. doi: 10.1021/sb500282v. [DOI] [PubMed] [Google Scholar]
- 46.Martinez A, Grabar TB, Shanmugam KT, Yomano LP, York SW, Ingram LO. Low salt medium for lactate and ethanol production by recombinant Escherichia coli B. Biotechnol Lett. 2007;29:397–404. doi: 10.1007/s10529-006-9252-y. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z, Kahn BB, Shi H, Xue BZ. Macrophage alpha1 AMP-activated protein kinase (alpha1AMPK) antagonizes fatty acid-induced inflammation through SIRT1. J Biol Chem. 2010;285:19051–19059. doi: 10.1074/jbc.M110.123620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ∆∆CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Kuhlman TE, Cox EC. Site-specific chromosomal integration of large synthetic constructs. Nucleic Acids Res. 2010;38:e92. doi: 10.1093/nar/gkp1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datas supporting the conclusions of this article are all available in the manuscript and supplementary.


