Figure 2.
KOSR, Serum, and 2i Support Unique Transcriptional States
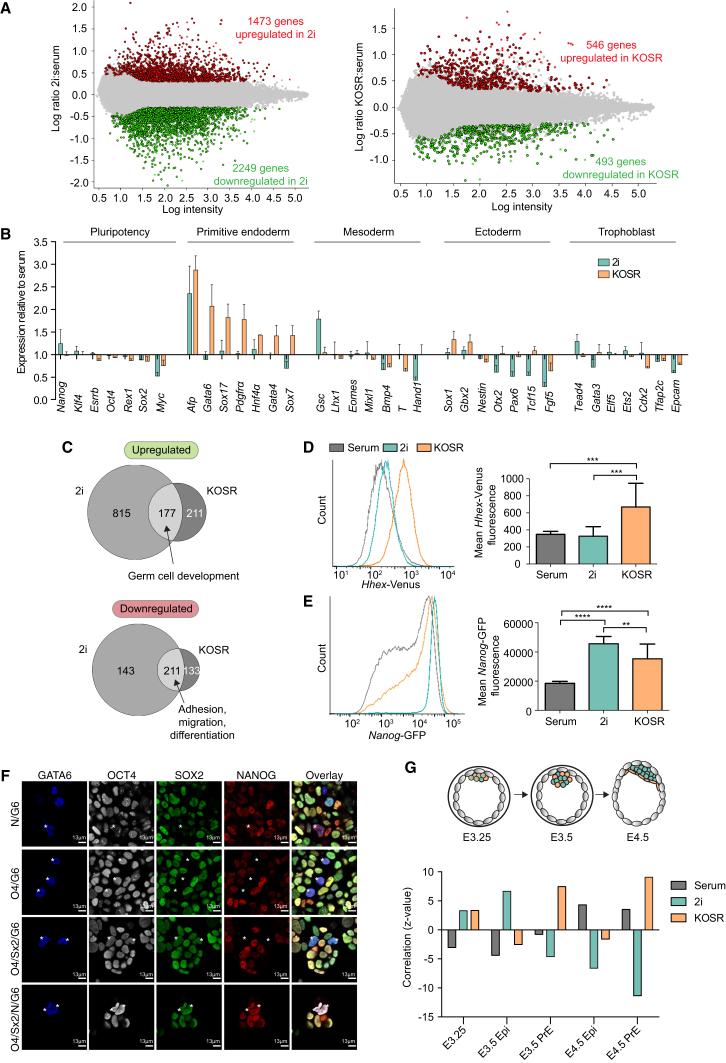
(A and B) Microarrays were carried out on 129/Ola ESCs cultured in serum, 2i, and KOSR conditions. At least three biological replicates of independent ESC lines were analyzed per condition. (A) Pairwise comparisons (FDR <0.05, ≥2-fold expression levels) were performed between serum-cultured and 2i- or KOSR-cultured ESCs to reveal non-redundant, significant changes in gene expression. Red dots represent significantly upregulated genes, green dots represent significantly downregulated genes, and gray dots represent genes that are not significantly changing in expression. (B) Expression changes in lineage markers in 2i- and KOSR-cultured ESCs relative to the expression in standard serum cultures.
(C) Venn diagrams illustrating the overlap between genes up- and downregulated in 2i and KOSR compared with standard serum cultures (identified in A).
(D and E) Flow cytometry profiles and corresponding mean fluorescence values for PECAM-1+ HV (D) or Nanog-GFP TNGB (E) ESCs cultured in serum, 2i, and KOSR. n = 3 independent experiments. Error bars indicate SD of the mean. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, Student's unpaired t test.
(F) Confocal optical section of ESCs cultured in serum, 2i, and KOSR immunostained for GATA6, OCT4, SOX2, and NANOG. Asterisks indicate coexpressing ESCs. The same observation was made in two independent cell lines.
(G) Graph showing the correlation of serum-, 2i-, and KOSR-cultured ESCs with published microarray data of different embryonic stages (Ohnishi et al., 2014).
See also Figure S2.