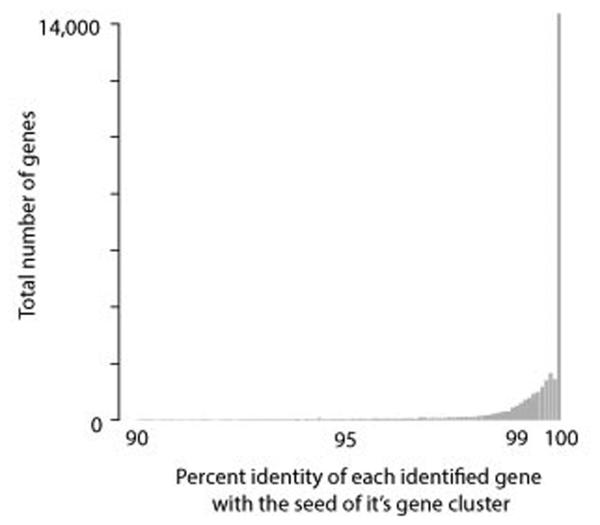
Extended Data Figure 9. Representative genes chosen for the final mobile gene dataset are highly similar to the genes that were filtered to reduce redundancy.
For each overlapping horizontally transferred region observed in cell-cell BLASTn comparisons between the reference genomes and single-cell assemblies, genes were clustered to identify unique genes and reduce the redundancy of the gene set. This step is essential for accurate abundance measurements of these genes in the metagenomic datasets after read alignment. All open reading frames from each overlapping horizontally transferred region were grouped using UCLUST. The nucleotide identities of each of the filtered genes and it’s the gene chosen for read alignment (i.e. the centroid) is plotted.