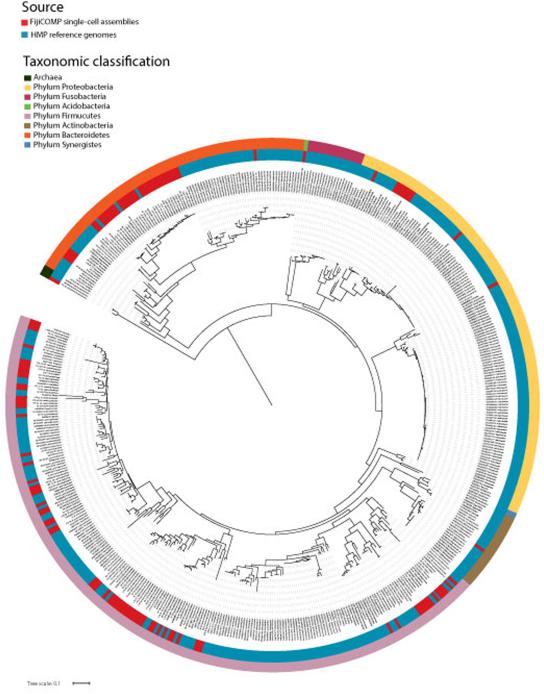
Extended Data Figure 1. Phylogeny of assemblies utilized in the study span the bacterial Tree of Life.
A phylogenetic tree constructed using a multiple sequence alignment of the full 16S rRNA gene or the V68 region of the 16S rRNA gene of all reference genomes and single-cell assemblies used in this analysis where available. 16S alignments were constructed using RDP. The tree was then assembled using FastTree. Support was low for all deep branches in the tree, therefore the archeal branch serves as the outgroup for illustrative purposes only. The outer color bar displays taxonomic associations for archea and bacterial phyla. The inner color bar displays the source of that operational taxonomic unit: HMP reference cells (blue) and FijiCOMP single cell assemblies (red). 16S rRNA gene sequences were not available for 70 FijiCOMP single-cell assembles, which are therefore not included in this tree.