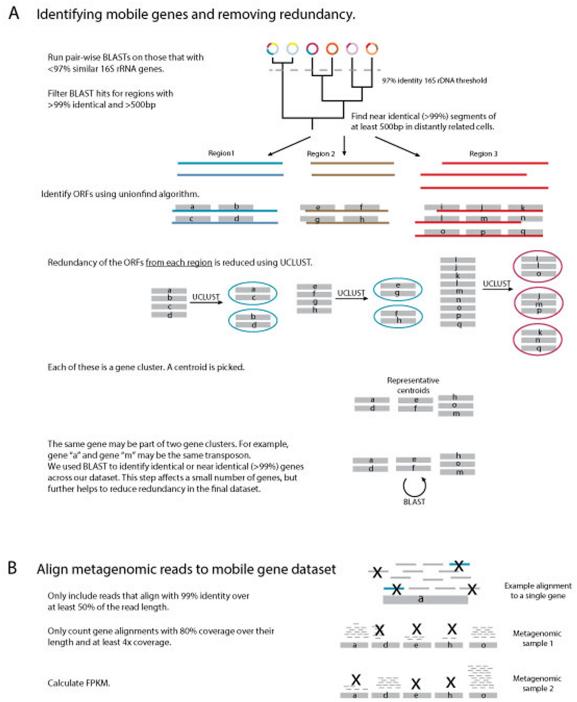
Extended Data Figure 2. Methodology for identifying horizontally transferred genes and assessing their distribution within the metagenomic samples.
Horizontally transferred regions were first identified using pair-wise BLASTs between HMP reference genomes and FijiCOMP single cell assemblies. Open reading frames were annotated within the horizontally transferred regions. Genetic redundancy was removed in the mobile gene set to ensure accurate abundance estimates using a combination of UCLUST and BLAST. Metagenomic reads were then aligned to the dataset of unique mobile genes. Alignments were filtered to retain only reads that aligned with 99% identity across over 50% of their read length. Abundances of genes in the metagenomic samples were determined for genes whose alignments had a minimum of 4x alignment depth over 80% of the gene length.