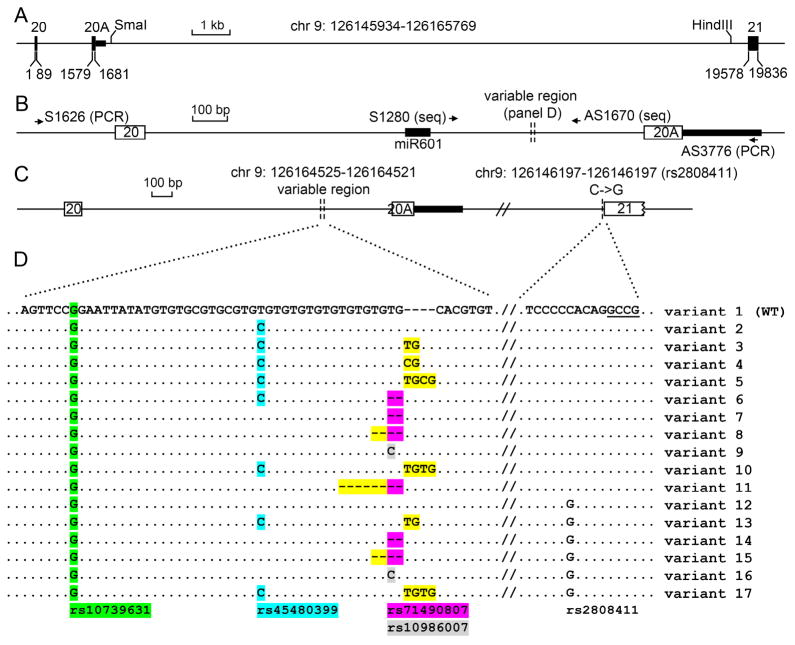
Fig 1. Diagram of the 3′ end of the DENND1A gene and splicing variants identified.
A. Scale diagram of ~20 kb spanning DENND1A exons 20 and 21; bases are numbered starting with the first base of exon 20 (chr 9:126145934) designated as base 1; exons are shown as boxes. The intronic regions between exons 20 and 20A and between exons 20A and 21 are 1490bp and 17897 bp, respectively. B. Magnified diagram of the region between exons 20 and 20A; note the difference in the scale bars in panels A and B. The hypervariable region between the vertical dotted lines was identified in PCR products generated with primers S1626 and AS3776; this hypervariable region is shown in detail in panel D. DNA sequencing (seq) of PCR products and cloned DNA was done using primers S1280 and AS1670. The 3′ end of miR601 (shown as a heavy black line) is 282 bp upstream of the hypervariable region. The heavy black line following exon 20A represents the DNA encoding the 3′ untranslated region of DENND1A V2 mRNA.
C. Locations of detected sequence variants. The hypervariable region was detected 330 bp upstream from exon 20A at chr 9: 126164525-126164521; and rs2808411 is adjacent to exon 21 at chr 9: 126146197-126146197.
D. Sequence variants in the hypervariable region. Known SNPs in this region are highlighted: green, rs10739631; cyan, rs45480399; purple, rs71490807; grey, rs10986007. rs2808411 (C->G) occurs 5 bp upstream of exon 21 (underlined). Newly identified variants at chr 9: 126164525-126164521 are highlighted in yellow.