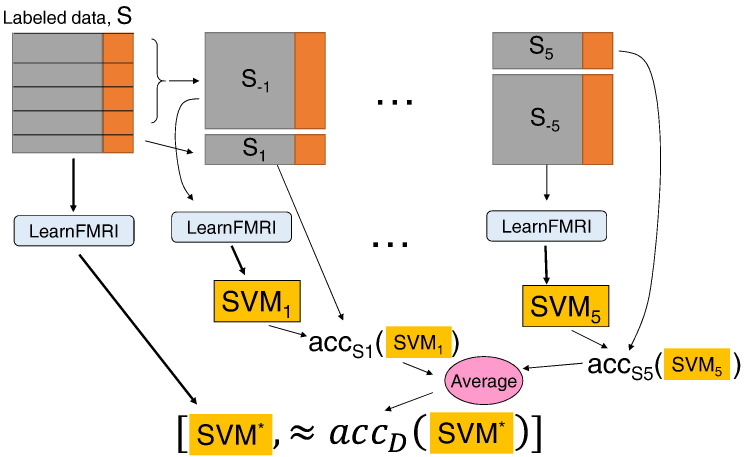
Fig. 2.
Illustration of the five-fold cross-validation procedure for evaluating the performance of running LearnFMRI on the labeled data S. This process first runs LearnFMRI on all of S, to produce the classifier SVM* – see left path. It then does 5 times more work, solely to estimate the actual performance of SVM* – i.e. how well SVM* will perform on unseen data, from the underlying distribution D. We denote the accuracy of SVM* on the underlying distribution D as accD(SVM*). This process divides S into 5 partitions. The procedure then runs LearnFMRI on 4/5 of the data (S1) to produce a classifier SVM1. It then evaluates this SVM1 on the remaining data (S − 1) – i.e. on the data that was not used to train SVM1. This produces the accuracy number accS1(SVM1). It does this 4 more times, on 4 other partitions [S − i, Si] of S, to produce 4 other estimates. We then use the average of these five {accSi(SVMi)} values as our estimate of SVM*'s accuracy.
Notice each of 5 “cross-validation” steps also requires running LearnFMRI, which note (from Fig. 1) has its own internal (4 fold) cross-validation steps, to find the best feature extractor and base learner. Hence, this involves “in fold” feature selection, etc.