Fig. (5).
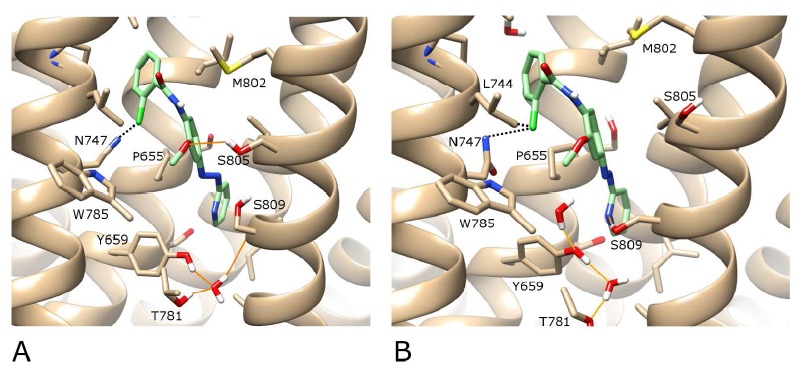
A) The best-ranked docking solution of the active isomer of alloswitch-1 (in green) in the complete wt structure of mGlu5 (in beige) where the ligand adopts a cis-amide conformation. B) After 100 ns of MD simulation “in the dark”. H-bonds are shown as orange lines and halogen interactions as dotted black lines. A protein-ligand H-bond between alloswitch-1 and S809 is formed during MD. For visualisation purposes, the backbone of TM6 is not shown, however the sidechains of residues W785 and T781 on TM6 are included (in the foreground). Regarding helix position, TM5 is foreground left, TM7 is foreground right. In the background, TM3 is centre-left and TM2 is centre-right.
