Fig. 1.
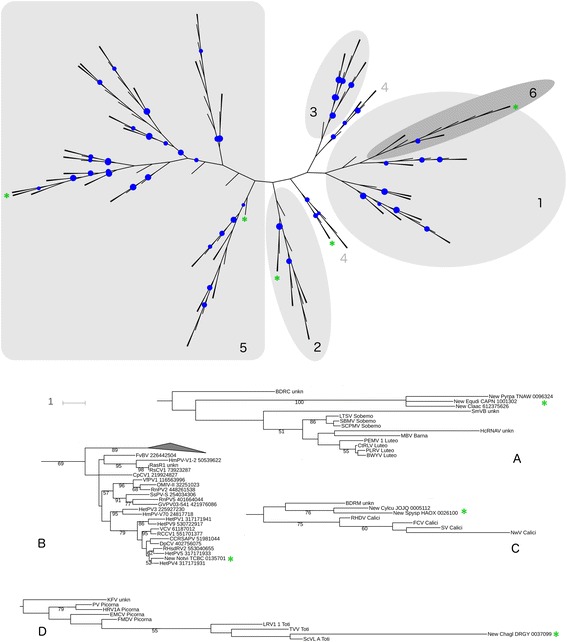
Maximum-likelihood tree of RNA-dependent RNA polymerases (RdRPs) from viruses of the picorna-like tribe. In the large tree (top), numbers indicate clades defined in Koonin et al. [34], and the bootstrap support is shown by indigo dots for all internal partitions where it was at least 60 %, with the dot size increasing as the values grow to 100 %. The branch lengths in the large tree are not to scale. Green asterisks indicate the position of new viruses. a–d The branch lengths of the subtrees are drawn to scale, with the bootstrap support shown for all partitions where it was higher than 50 %. Novel viruses are marked by green asterisks; their identifiers start with “New” and are followed by the ID of the 1000 Plants Genomes Project scaffold. Virus names, short descriptions of taxonomic position, and GenBank ID numbers are taken from Koonin et al. [34] and Nibert et al. [33], except that all sequence identifiers in the latter source referred to the nucleotide sequences and were replaced in this study by the IDs of their protein products that were actually aligned. a A clade of virus RdRPs from diverse hosts is a deep branch within the picornavirus-like Clade 2. b New alphapartitivirus from Nothoceros vincentianus (left-most branch in Clade 5 on top). c Two sequences from Zygnematales may be distantly related to picornavirus-like Clade 4. d A totivirus from Chaetosphaeridium globosum
