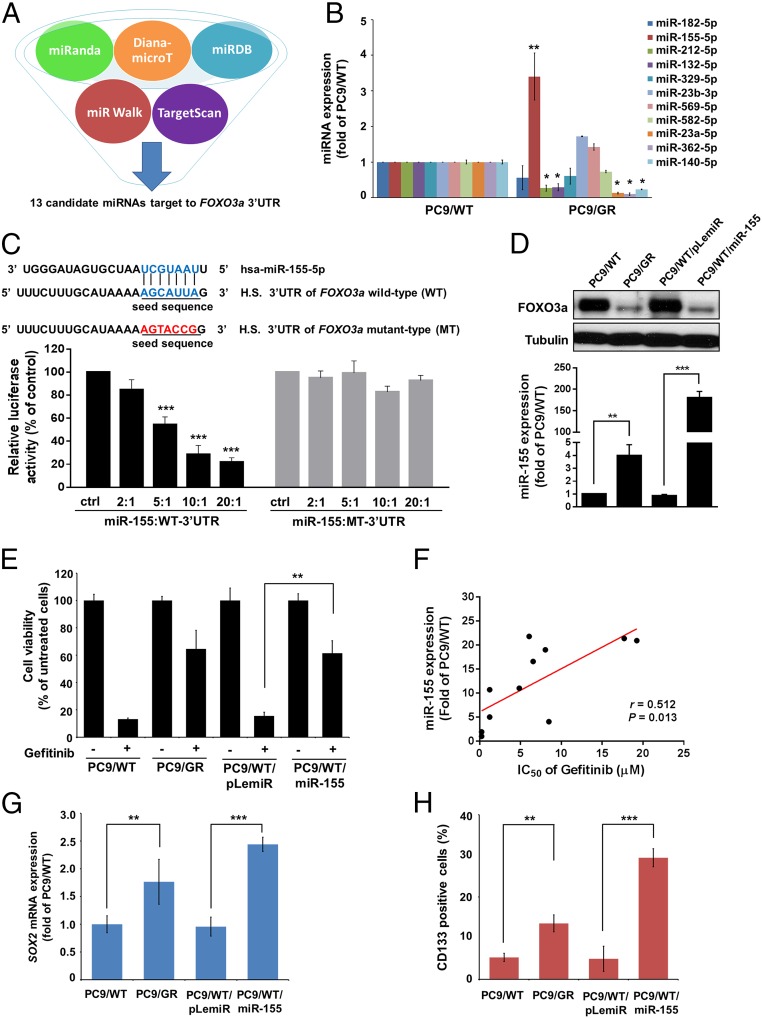
Fig. 3.
miR-155 is involved in FOXO3a-mediated gefitinib sensitivity and CSC characteristics. (A) Schematic of the bioinformatic analyses of candidate microRNAs that targeted FOXO3a. The putative microRNAs targeting FOXO3a in miRWalk, TargetScan, miRanda, miRDB, and Diana-microT were searched and 13 candidate microRNAs were filtered out for real-time RT-PCR analysis. (B) Real-time RT-PCR analysis of the differential expression of the 11 microRNAs in PC9/WT compared with PC9/GR cells. (C) A schematic diagram representing the predicted miR-155 binding sequences or mutated versions (Upper). Luciferase activity (Lower) of wild-type FOXO3a-3′UTR (WT-3′UTR) and mutant-type FOXO3a-3′UTR (MT-3′UTR) reporter genes were measured using the Dual-Luciferase Reporter Assay System in PC9/WT cells transfected with miR-155 at different ratios. (D) The effects of miR-155 overexpression on endogenous FOXO3a protein expression in PC9/WT cells. FOXO3a and miR-155 expression were measured by Western blot analysis and real-time RT-PCR, respectively. Tubulin was used as a loading control. (E) The functions of miR-155 in regulating gefitinib sensitivity were assayed by comparing PC9/WT/pLemiR and PC9/WT/miR-155 cells that were treated with or without 4 μM gefitinib. Cell viability was determined by MTT assay. (F) The correlation between miR-155 expression and the IC50 of gefitinib with doses for the lung cancer cell lines. (r = 0.512; P = 0.013; Pearson’s correlation coefficient). Expression of miR-155 affected (G) SOX2 mRNA levels by real-time RT-PCR and (H) CD133+ cells by flow cytometric analysis in PC/WT cells with the expression of pLemiR-155 and control vector. The results are shown as the means ± SD of three independent experiments, each performed in triplicate. *P < 0.05, **P < 0.01, and ***P < 0.001 (two-tailed Student's t test).