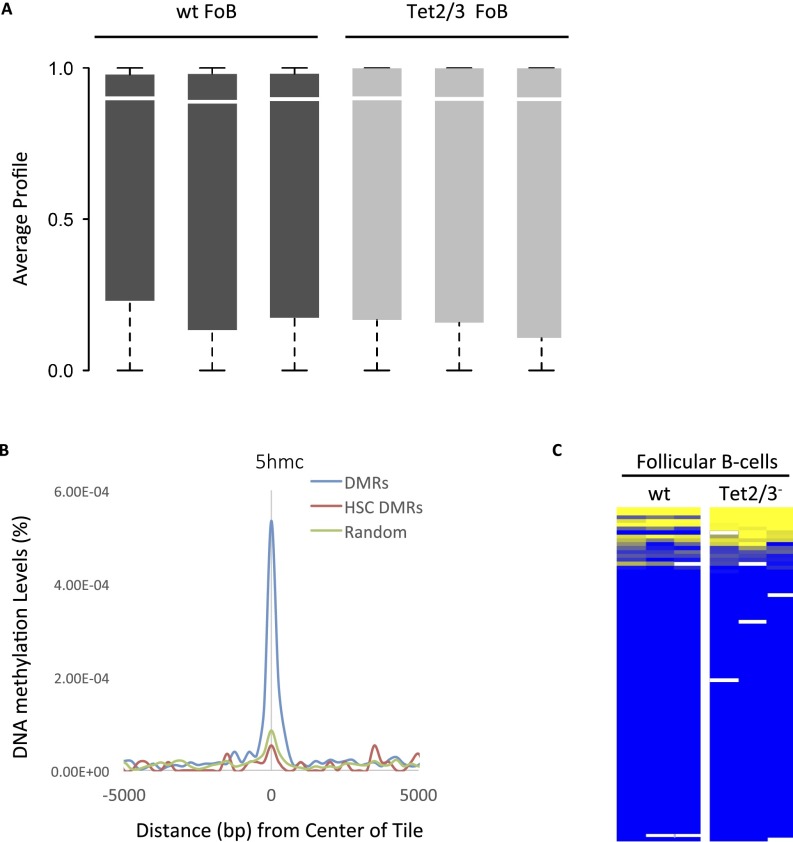
Fig. S1.
The 5mC and 5hmC methylation distribution. (A) Box plots of global RRBS methylation values for all replicates of wild-type and Tet2/3− follicular B-cell samples. (B) Enrichment of 5hmc in hematopoietic progenitor cells (GSE65895) as a function of distance from the center of each DMR (Fig. 1), compared with a random control. Tiles specifically unmethylated in HSCs are not enriched. The 5hmC levels were determined by HELP-GT (HpaII tiny fragment enrichment by ligation-mediated PCR with β-glucosyl transferase) in Tel1+/+ as opposed to Tet1−/− hematopoietic progenitor cells (70). Average coverage was 10–15× per base. (C) DNA methylation levels of the promoter regions for genes that harbor differentially methylated tiles (n = 356). Only 11 (3%) show a significant difference between wt and DKO. Yellow represents high and blue represents low DNA methylation levels.