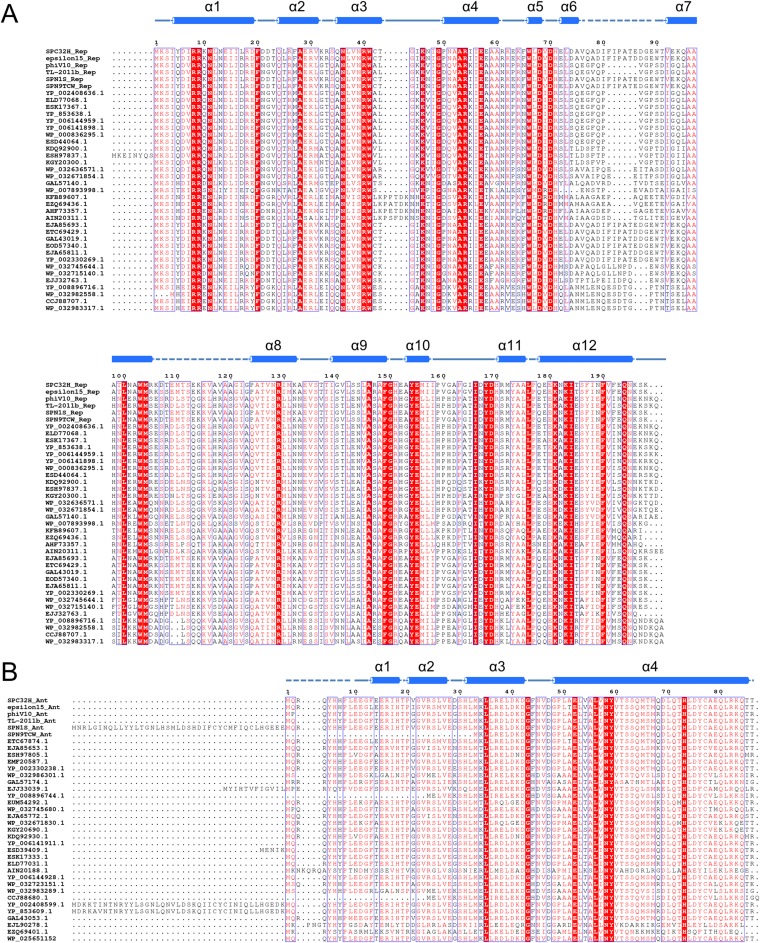
Fig. S2.
Sequence alignments of Rep (A) and Ant (B). Multialignment of S. Typhimurium Rep (UniProtKB accession no. T1S9Z0) against Rep from Salmonella phage epsilon15 (UniProtKB accession no. Q858D7), Escherichia phage phiV10 (UniProtKB accession no. Q286X7), Escherichia phage TL-2011b (UniProtKB accession no. G9L6A6), Salmonella phage SPN1S (UniProtKB accession no. H2D0H9), Salmonella phage SPN9TCW (UniProtKB accession no. M1F232), E. coli O118:H16 str. 2009C-4446 (UniProtKB accession no. A0A028E2H1), E. coli KTE235 (Ensembl Bacteria accession no. ELD77068), E. coli UMEA 3290-1 (Ensembl Bacteria accession no. ESK17367), E. coli MS 196-1 (UniProtKB accession no. D8BV09), E. coli O7:K1 str. CE10 (RefSeq accession no. YP_006144959), E. coli NA114 (RefSeq accession no. YP_006141898), E. coli MS 185-1 (RefSeq accession no. WP_000836295), E. coli 908519 (UniProtKB accession no. V0VMF3), Salmonella enterica subsp. enterica serovar Bareilly str. CFSAN000179 (EMBL-WGS accession no. KDQ92900), S. enterica subsp. enterica serovar Agona str. 632182-2 (Ensembl Bacteria accession no. ESH97837), Citrobacter koseri (UniProtKB accession no. A0A0A5IUU2), Enterobacter sp. MGH 15 (RefSeq accession no. WP_032636571), Enterobacter cloacae (RefSeq accession no. WP_032671854), Escherichia vulneris NBRC 102420 (UniProtKB accession no. A0A090UX00), Pantoea sp. GM01 (UniProtKB accession no. J2L6Z7), Serratia grimesii (UniProtKB accession no. A0A084YWL3), Serratia marcescens BIDMC 80 (Ensembl Bacteria accession no. EZQ69436), Candidatus Sodalis pierantonio str. SOPE (UniProtKB accession no. W0HLH1), Yersinia kristensenii (UniProtKB accession no. A0A088L5A4), S. enterica subsp. enterica serovar Newport str. CVM 19470 (Ensembl Bacteria accession no. EJA85693), S. enterica subsp. enterica serovar Enteritidis str. 3402 (UniProtKB accession no. V7Y6W8), Citrobacter werkmanii NBRC 105721 (UniProtKB accession no. A0A090TWG5), Citrobacter freundii GTC 09629 (Ensembl Bacteria accession no. EOD57340), S. enterica subsp. enterica serovar Newport str. CVM 19443 (Ensembl Bacteria accession no. EJA65811), E. coli O127:H6 (strain E2348/69/EPEC) (UniProtKB accession no. B7UGT4), Klebsiella oxytoca (RefSeq accession no. WP_032745644), Enterobacter aerogenes (RefSeq accession no. WP_032715140), Klebsiella pneumoniae (EMBL CDS accession no. AHM80771), Cronobacter sakazakii CMCC 45402 (UniProtKB accession no. V5TYG0), Cronobacter malonaticus (RefSeq accession no. WP_032982558), Cronobacter turicensis 564 (UniProtKB accession no. K8BK39), and C. malonaticus (RefSeq accession no. WP_032983317). Multialignment of Ant (UniProtKB accession no. T1SA45) against Ant from Salmonella phage epsilon15 (UniProtKB accession no. Q858F6), Escherichia phage phiV10 (UniProtKB accession no. Q286Z4), Escherichia phage TL-2011b (UniProtKB accession no. G9L6E3), Salmonella phage SPN1S (UniProtKB accession no. H2D0F7), Salmonella phage SPN9TCW (UniProtKB accession no. M1EZ64), S. enterica subsp. enterica serovar Enteritidis str. 3402 (UniProtKB accession no. V7Y2G6), S. enterica subsp. enterica serovar Newport str. CVM 19470 (Ensembl Bacteria accession no. EJA85653), S. enterica subsp. enterica serovar Agona str. 632182-2 (Ensembl Bacteria accession no. ESH97805), C. freundii GTC 09479 (EMBL-WGS accession no. EMF20587), E. coli O127:H6 (UniProtKB accession no. B7UGQ3), C. malonaticus (RefSeq accession no. WP_032986301), E. vulneris NBRC 102420 (UniProtKB accession no. A0A090V1M3), K. pneumoniae subsp. pneumoniae KPNIH10 (UniProtKB accession no. A0A0E1DF28), C. sakazakii CMCC 45402 (UniProtKB accession no. V5U0K5), Enterobacter sp. MGH 15 (Ensembl Bacteria accession no. EUM54292), K. oxytoca (RefSeq accession no. WP_032745680), S. enterica subsp. enterica serovar Newport str. CVM 19443 (Ensembl Bacteria accession no. EJA65772), E. cloacae (RefSeq accession no. WP_032671830), C. koseri (UniProtKB accession no. A0A0A5IVX8), S. enterica subsp. enterica serovar Bareilly str. CFSAN000179 (Ensembl Bacteria accession no. KDQ92930), E. coli NA114 (RefSeq accession no. YP_006141911), E. coli 908519 (UniProtKB accession no. V0V944), E. coli UMEA 3290-1 (EMBL-WGS accession no. ESK17333), E. coli KTE235 (EMBL-WGS accession no. ELD77031), Y. kristensenii (UniProtKB accession no. A0A088L842), E. coli O7:K1 str. CE10 (RefSeq accession no. YP_006144928), E. aerogenes UCI 15 (RefSeq accession no. WP_032723151), C. malonaticus (RefSeq accession no. WP_032983289), C. turicensis 564 (UniProtKB accession no. K8BRJ5), E. coli O7:K1 (strain IAI39/ExPEC) (RefSeq accession no. YP_002408599), E. coli O1:K1/APEC (RefSeq accession no. YP_853609), C. werkmanii NBRC 105721 (UniProtKB accession no. A0A090TUF1), Pantoea sp. GM01 (UniProtKB accession no. J2LY07), S. marcescens BIDMC 80 (Ensembl Bacteria accession no. EZQ69401), and E. coli FCP1 (RefSeq accession no. WP_025651152). Secondary structure elements were assigned by PyMOL (The PyMOL Molecular Graphics System, www.pymol.org). Every tenth residue is marked by a black dot. Strictly 100% conserved residues are highlighted in red. Cylinders above the sequences denote α-helices. A dotted line denotes disordered regions.