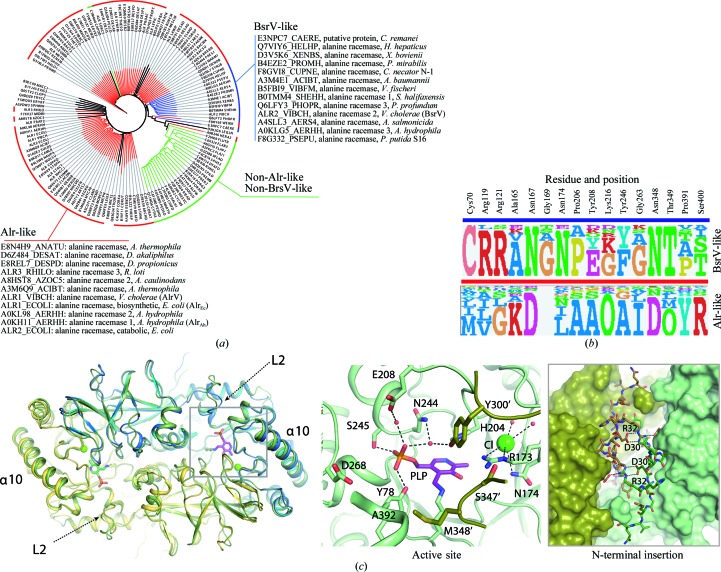
Figure 4.
The identification of 16 residues that constitute the molecular footprint for BsrV multispecificity and experimental validation. (a) Tree of BsrV orthologues obtained through bioinformatic sequence analysis (see §2). Alr-like enzymes are highlighted in red, Bsr-like enzymes in blue and non-Alr-like, non-Bsr-like enzymes in green; 15 outlier proteins that do not group with any of these families are shown in black. All Bsr-like family members (including BsrV) and a small subset of Alr-like enzymes (including AlrV and AlrEc) are listed. (b) Molecular footprint. Sequence composition (displayed as sequence logos) of the specificity-determining positions for Alr/BsrV-like racemases. The specific residues and their positions are listed at the top. (c) Structural analysis of the broad-spectrum racemase from A. hydrophila (BsrAh). A structural superimposition of BsrV with BsrAh is shown on the left. Loop L2 and the α-helix α10 at the entry channel are labelled. The central panel shows a close-up view of the BsrAh catalytic site. Relevant residues and PLP are shown as sticks; water molecules and Cl− ions are shown as red and green spheres, respectively. In the right box, a close-up view of the back-side region (180° rotation view) of BsrAh with the N-terminal insertion in stick representation is shown. Polar interactions between relevant residues of the N-terminal extensions from both monomers are represented as dotted lines. See also Supplementary Fig. S4.