Abstract
DNA methylation plays an important role in gene expression and virulence in some pathogenic bacteria. In this report, we describe DNA methyltransferases (MTases) present in human pathogenic bacteria and compared them with related species, which are not pathogenic or less pathogenic, based in comparative genomics. We performed a search in the KEGG database of the KEGG database orthology groups associated with adenine and cytosine DNA MTase activities (EC: 2.1.1.37, EC: 2.1.1.113 and EC: 2.1.1.72) in 37 human pathogenic species and 18 non/less pathogenic relatives and performed comparisons of the number of these MTases sequences according to their genome size, the DNA MTase type and with their non-less pathogenic relatives. We observed that Helicobacter pylori and Neisseria spp. presented the highest number of MTases while ten different species did not present a predicted DNA MTase. We also detected a significant increase of adenine MTases over cytosine MTases (2.19 vs. 1.06, respectively, p < 0.001). Adenine MTases were the only MTases associated with restriction modification systems and DNA MTases associated with type I restriction modification systems were more numerous than those associated with type III restriction modification systems (0.84 vs. 0.17, p < 0.001); additionally, there was no correlation with the genome size and the total number of DNA MTases, indicating that the number of DNA MTases is related to the particular evolution and lifestyle of specific species, regulating the expression of virulence genes in some pathogenic bacteria.
Electronic supplementary material
The online version of this article (doi:10.1007/s12088-015-0562-4) contains supplementary material, which is available to authorized users.
Keywords: DNA methyltransferases, Comparative genomics, Pathogenic bacteria
Introduction
DNA methylation plays a critical role in epigenetic gene regulation in prokaryotes and eukaryotes. In bacteria, DNA methylation functions through the regulation of gene expression that helps bacteria to cope with environmental changes, including the nutrient availability, temperature, pH and osmolarity [1, 2]. Important roles in the biology of bacteria are influenced by epigenetic mechanisms, including the timing of DNA replication, the partitioning of nascent chromosomes into daughter cells, the repair of DNA and the timing of the transposition and conjugal transfer of plasmids [3]. DNA methylation in bacteria occurs at the C-5 or N-4 positions of cytosine and at the N-6 position of adenine, and it is catalyzed by DNA methyltransferases (MTases) [2, 4]. DNA MTases in bacteria have been classified into two primary groups, one of which is associated with restriction modification systems (R–M systems) and the other with orphan or solitary MTases, which are not associated with a restriction enzyme. The R–M systems are in turn classified into three types (types I through III) and act as an immune system equivalent for preventing the invasion of foreign DNA by producing a double-strand break in specific unmethylated DNA sequences; this break is impeded in the host genome by the proper methylation of these sequences by the DNA MTases in the R–M systems [2, 5]. The orphan MTases, which are believed to have evolved from R–M systems [6], comprise DNA adenine MTase (Dam), cell cycle-regulated MTase (CcrM) and DNA cytosine MTase (Dcm) [2, 3]. Dam MTase has been shown to be related to the expression of virulence genes in pathogenic bacteria, including Haemophilus influenzae, Salmonella enterica, Yersinia spp. and Vibrio cholerae, among others [2, 4], and an association between MTases of the type III R-M system is also involved in the expression of virulence genes in Neisseria gonorrhoeae [7]. Considering that DNA MTases can be associated with virulence in pathogenic bacteria and that complete reports about DNA MTases in primary human pathogenic bacteria have not been presented, we decided to identify the DNA MTases in human pathogenic bacteria by using comparative genomics to understand their distribution in a way that could offer knowledge with respect to their functionality. Additionally, we performed a comparison with respect to non-/less pathogenic relatives.
Materials and Methods
By drawing from the KEGG database [8], we included 37 different species of human pathogenic bacteria that represent the primary causes of human bacterial infections. We excluded most opportunistic pathogens with the exception of Streptococcus pneumoniae and Neisseria meningitidis, which are important causes of human infections in specific age groups. When more than two bacteria with the same phenotype were available, we included only one (the most common). We additionally included 18 phylogenetically related non-/less pathogenic bacteria (according to the description in the NCBI database of non- or rarely pathogenic). These bacteria were obtained according to the availability of phylogenetically related (from the same genus) and completely sequenced bacteria and were included as a reference for the related pathogenic bacteria. In 40 of the 55 species, we included 2–4 subspecies from each species (according to the availability of genomes in the KEGG database) and obtained an average for the number of analyzed MTases. When possible, these subspecies were chosen according to their higher genome representativeness, as identified in the genome group’s report in the NCBI. We were not able to include more subspecies for the other 15 species because of the lack of other genomes in the KEGG database.
By using the PYTHON programming language, we identified the enzymatic numbers in the KEGG database associated with cytosine MTase activity, EC: 2.1.1.37 (C5-cytosine) and EC: 2.1.1.113 (cytosine-N4-specific), and with N6-adenine MTase activity, EC: 2.1.1.72; we obtained all the KO (KEGG orthology) numbers associated with these enzymatic numbers in all the analyzed bacteria. We also obtained the genome size and the number of plasmids of each bacterium from the NCBI database.
In order to identify the conserved residues and active sites we generated a consensus sequence for each KO, first we obtained and filtered the sequences at the 60 % of identity and then, by using the cons program from EMBOSS system we obtained the consensus sequences and posteriorly, we aligned them versus a superfamily model from where we obtained the catalytic amino acids.
Statistical Analysis
Our descriptive statistics consisted of the frequencies and percentages for qualitative variables, and we used the means, medians and ranges for quantitative variables. Pearson’s correlation test was used to associate two quantitative variables with parametric distributions, whereas a Spearman correlation test was used for non-parametric distributions, yielding a correlation coefficient in both cases (r). The statistical significance was set at ≤0.05, and the statistical analysis was performed with SPSS v. 10.0.
Results
Identification of the KEGG Orthology (KOs) Groups
For the cytosine-C5 MTase enzymatic activity (EC: 2.1.1.37), we found the KO K00558 (Dcm, DNMT1, DNA cytosine-5 MTase 1); for the cytosine-N4-specific MTase activity (EC: 2.1.1.113), we found the KO K00590 (site-specific DNA MTase cytosine-N4-specific); and for the N6-adenine-specific MTase activity (EC: 2.1.1.72), we found the following KOs: K00571 (site-specific DNA MTase), K03427 (HsdM, type I restriction enzyme M protein), K06223 (Dam, DNA adenine methylase), K07316 (mod, adenine-specific DNA MTase), K07317 (adenine-specific DNA MTase), K07318 (adenine-specific DNA MTase), K07319 (yhdJ, adenine-specific DNA MTase) and K13581 (ccrM, modification methylase). The consensus sequences with their respective catalytic amino acids are presented in Supplementary Figure.
Grouping the Variables
To analyze the data, we grouped the KOs associated with cytosine MTase activity (K00558 and K00590), with adenine MTase activity (K00571, K03427, K06223, K07316, K07317, K07318, K07319 and K13581) and with adenine MTase activity not associated with restriction modification systems (K06223, K07317 and K13581). Considering that the MTases associated with a type II restriction modification system were only sporadically found in K00571, K07318 and K07319, these KOs were not grouped. The following KOs were also analyzed separately: K06223 (Dam methylase), K00558 (Dcm methylase), K00590 (site-specific DNA MTase cytosine-N4-specific), K03427 (HsdM, type I restriction enzyme M protein) and K07316 (mod, associated with the type III restriction modification system).
Descriptive Results
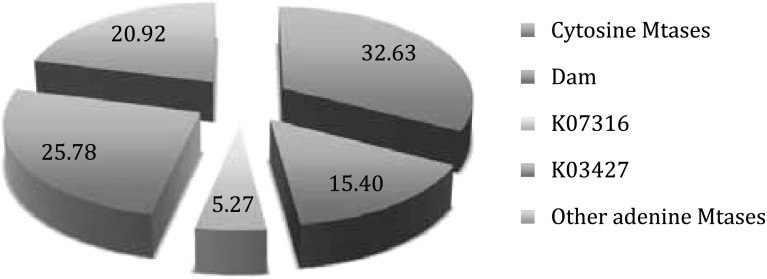
The general distribution was expressed as percentages of the primary DNA MTase types, and it is represented in Fig. 1. We can observe that most DNA MTases belong to adenine MTases, approximately one-third are cytosine MTases and another third are adenine MTases associated with type I or type III restriction modification systems.
Fig. 1.
Percentages of primary DNA MTase types of the total MTase number found in the 55 analyzed bacteria
Qualitative Expression
When the results were analyzed as qualitative variables, they indicated the presence or absence of a specific MTase homolog in at least one subspecies of each species from the 55 analyzed bacteria. We observed that 45/55 bacteria (81.81 %) presented at least one DNA MTase, 43/55 bacteria (78.18 %) presented at least one adenine MTase, and 24/55 (43.63 %) bacteria presented at least one Dam MTase. Twenty-nine of the fifty-five bacteria (52.72 %) presented at least one cytosine MTase, and among these, 28/29 (96.55 %) presented at least one Dcm MTase (K00558). Only 3/55 (5.45 %) bacteria presented at least one N4-cytosine MTase. Thirty-four of the fifty-five bacteria (61.81 %) presented at least one homolog of the HsdM, type I restriction enzyme M protein (K03427), 11/55 (20 %) bacteria presented at least one homolog of the mod MTase associated with the type III restriction modification system (K07316), and 25/55 bacteria (45.45 %) presented at least one adenine MTase not associated with a restriction modification system. Of these MTases, only 2/25 (8 %), were predicted to be CcrM MTases, with one in Brucella melitensis and the other in Helicobacter pylori. The highest total number of DNA MTases was found in Neisseria lactamica bacteria, followed by H. pylori and N. gonorrhoeae, and no MTase was found in 10 bacteria belonging to the genera Bordetella, Coxiella, Chlamydia, Mycobacterium, Ehrlichia, Rickettsia and Leptospira.
From all the analyzed bacteria, only 21/55 (38.18 %) species had at least one subspecies with a plasmid (6/21 species had at least one subspecies with more than one plasmid), and from these, only 6/21 (28.57 %) had at least one subspecies with a DNA MTase encoded in a plasmid. Additionally, we observed that 13 out of the global 178.40 (this number is the sum of the total DNA MTases in all the bacteria) DNA MTases (7.29 %) were encoded in a plasmid, from these 13 MTases, 9 belonged to K07319, two to K00558 and two more to K06223.
Quantitative Expression
When the descriptive results of the 55 analyzed bacteria were expressed in numbers (as quantitative variables), we observed that the average (median) (range) of the global number of DNA MTases was 3.24 (2.33) (0–14); for the cytosine MTases, it was 1.06 (0.33) (0–8.5), and for adenine MTases, it was 2.19 (2.00) (0–10). The K03427 presented an average (median) (range) of 0.84 (0.75) (0–3), and the K07316 of 0.17 (0) (0–2); finally, the average (median) (range) of the adenine MTases not associated with a restriction modification system was 0.56 (0) (0–5).
Description of Intra-species Variability
To observe the variability in the number of DNA MTases analyzed within the 33 species that included more than one subspecies and had at least 1 MTase, we obtained the SD within each species and afterward calculated the mean and ranges of these values. The mean (median) (range) of these values were 1.16 (0.58) (0–5.29).
Comparative Results
Correlation of MTases and the Genome Size
When we associated the genome size with the total number of DNA MTases, we could not observe a significant correlation, with r = 0.161 and p = 0.239, but when the correlation was performed with specific types of DNA MTases, we only observed a significant correlation with the number of cytosine MTases with r = 0.300 and p = 0.026.
Comparing the Numbers of the Different DNA MTase Types
When we compared the number of DNA MTases among the different types, we observed a significant increase in adenine with respect to cytosine MTases, and the average (median) of 2.19 (2.00), compared with 1.06 (0.33), p < 0.001. There was also a significant increase in K03427 (MTases associated with the type I restriction modification system) with respect to K07316 (MTases associated with the type III restriction modification system), and the average (median) was 0.84 (0.75), compared with 0.17 (0), p < 0.001.
Discussion
Unlike prokaryotic DNA methylation, the DNA methylation in eukaryotes occurs only in cytosine-C5 (EC: 2.1.1.37), which is performed by three different DNA MTases, including DNMT1 (K00558), that methylates hemymethylated DNA and therefore provides heritability of epigenetic information, and DNMT3A (K17398) and DNMT3B (K17399), which are also named de novo MTases and set up new methylation patterns [9]. In bacteria the analysis of restriction endonuclease sites can be used for the rapid identification of specific species in different pathogenic organisms [10–12] and for a better understanding of their evolution [13].
In the present report, we identified the DNA MTases found in primary human pathogenic bacteria and their phylogenetically related non-/less pathogenic bacteria on the basis of comparative genomics according to the KEGG database. REBASE database [14] was not used considering the lack of organization and specificity of its DNA MTases classification. To date, although some reports related to DNA MTases have been published about specific pathogenic bacteria [15–17], no reports have summarized the identification of these DNA MTases in the primary human pathogenic bacteria.
Among all the analyzed bacteria, we did not find any predicted DNA MTases in the following 10 different species: Coxiella burnetii, Chlamydia pneumoniae, Chlamydia trachomatis, Ehrlichia chaffeensis, Mycobacterium leprae, Rickettsia prowazekii, Rickettsia peaccoki, Bordetella pertussis, Mycobacterium indicus pranii and Leptospira biflexa; of these species, seven were pathogenic and three were non-/less pathogenic (see Table 1). It is interesting that 5 of the 7 pathogenic species, namely, C. burnetii, C. pneumoniae, C. trachomatis, E. chaffeensis and M. leprae, presented reduced genomes as well as obligate intracellular lifestyles. According to the literature, we were unable to detect reported DNA MTases in the bacterial genera Bordetella, Coxiella and Ehrlichia. Nevertheless, in the case of C. trachomatis, an absence of Dam methylation was reported, but low levels of Dcm methylation are found in its genome [18], and some subspecies of C. trachomatis and C. pneumoniae presented adenine MTases but not cytosine MTases in the NCBI/gene database, indicating a lack of concordance between the NCBI database and the literature reports. Additionally, a functional Mycobacterium spp. adenine MTase (MamA) associated with gene expression and survival under hypoxic conditions has recently been reported in M. tuberculosis, and this MTase has also been identified in M. tuberculosis relatives, including M. leprae [16]. However, according to our results, an absence of the classical Dam or Dcm methylation of their specific recognition sites in Mycobacteria spp. has been shown [19]. However, with a search of homologs (Blast-p with 60 % of sequence coverage and E-value ≤10−3) of the MamA sequence in 2757 bacterial and archeal genomes from the NCBI database we found MamA homologs in 639 different organisms including 11 of the bacteria analyzed: Campylobacter jejuni, Corynebacterium diphteriae, Francisella novicida, H. influenzae, Helicobacter cinaedi, H. pylori, Leptospira interrogans, M. leprae, M. tuberculosis, Nocardia brasiliensis and Treponema primitia, which indicates that unidentified MTases can also have more homologs in the bacteria analyzed. The complete list of the organisms with a MamA homolog is presented in the Supplementary Table.
Table 1.
Number of specific DNA methyltransferases in the 55 analyzed bacteria
| Bacteria (KEGG genome abbreviation) | Total MTases | Adenine MTases | Cytosine MTases | Dcm | Dam | K07316 | K03427 |
|---|---|---|---|---|---|---|---|
| Bacillus anthracis (ban, bar, bat) | 1.33 | 1.00 | 0.33 | 0.33 | 0.00 | 0.00 | 0.00 |
| Bacillus subtilis subsp. subtilis 168 (bsu)a | 4.00 | 1.67 | 2.33 | 2.33 | 0.00 | 0.00 | 0.67 |
| Bordetella pertussis (bpe, bpc, bper) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Bordetella petrii ASM6720v1 (bpt)a | 9.00 | 3.00 | 6.00 | 6.00 | 0.00 | 1.00 | 1.00 |
| Borrelia burgdorferi (bbu, bbz, bbn) | 0.33 | 0.00 | 0.33 | 0.33 | 0.00 | 0.00 | 0.00 |
| Brucella melitensis (bme, bmi, bmz) | 2.00 | 2.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| Campylobacter hominis ATCC BAA-381 (cha)a | 3.00 | 2.00 | 1.00 | 1.00 | 0.00 | 1.00 | 0.00 |
| Campylobacter jejuni (cje, cji, cju) | 2.33 | 2.00 | 0.33 | 0.33 | 0.00 | 0.00 | 1.00 |
| Chlamydiae pneumoniae (cpn, cpa, cpj) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Chlamydiae trachomatis (ctr, ctb, ctk) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Clostridium botulinum (cbo, cba, cbh) | 3.67 | 1.00 | 2.67 | 2.67 | 0.00 | 0.00 | 1.00 |
| Clostridium tetani (ctc, ctet) | 1.50 | 1.50 | 0.00 | 0.00 | 0.50 | 0.00 | 0.50 |
| Corynebacterium diphtheriae (cde, cdi, cdp, cdh) | 2.50 | 2.50 | 0.00 | 0.00 | 0.00 | 0.25 | 2.25 |
| Corynebacterium glutamicum (cgb, cgu, cgt)a | 1.33 | 0.33 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 |
| Coxiella burnetii (cbu, cbs, cbd) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Ehrlichia chaffeensis (ech, cha, echj) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Escherichia coli K12 (eco, ecj, ecd)a | 7.33 | 5.67 | 1.67 | 1.67 | 1.00 | 0.00 | 1.00 |
| Escherichia coli O:157 (ecs, ece, ecf) | 3.67 | 2.67 | 1.00 | 1.00 | 1.00 | 0.00 | 0.67 |
| Francisella novicida U112 (ftn)a | 3.00 | 3.00 | 0.00 | 0.00 | 0.00 | 1.00 | 2.00 |
| Francisella tularensis (ftu, ftf, ftr) | 1.33 | 1.33 | 0.00 | 0.00 | 0.00 | 0.33 | 1.00 |
| Haemophilus ducreyi (hdu) | 1.00 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| Haemophilus influenzae (hin, hit, hip) | 5.67 | 4.00 | 1.67 | 1.67 | 0.67 | 0.00 | 2.33 |
| Haemophilus parainfluenzae T3T1 (hpr)a | 2.00 | 2.00 | 0.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| Helicobacter cinaedi (hcp, hcb)a | 7.50 | 4.50 | 3.00 | 3.00 | 1.00 | 1.00 | 0.50 |
| Helicobacter pylori (hpy, hpj, hpd) | 13.00 | 9.67 | 3.33 | 3.00 | 0.33 | 0.33 | 3.00 |
| Legionella pneumophila (lpn, lph, lpo) | 1.67 | 1.33 | 0.33 | 0.33 | 1.33 | 0.00 | 0.00 |
| Leptospira biflexa (lbf, lbi)a | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Leptospira interrogans (lil, lie, lic) | 1.33 | 1.00 | 0.33 | 0.00 | 0.00 | 0.00 | 1.00 |
| Listeria innocua Clip11262 (lin)a | 3.00 | 3.00 | 0.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| Listeria monocytogenes (lmo, lmf, lmt) | 2.33 | 2.00 | 0.33 | 0.33 | 0.67 | 0.00 | 0.33 |
| Mycobacterium indicus pranii MTCC 9506 (mip)a | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Mycobacterium leprae (mle, mlb) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Mycobacterium tuberculosis (mtu, mtul, mtv) | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| Mycoplasma genitalium (mge, mgu, mgc) | 0.33 | 0.33 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Mycoplasma hominis ATCC23114 (mho)a | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| Mycoplasma pneumoniae (mpn, mpj, mpm) | 2.00 | 2.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| Neisseria gonorrhoeae (ngo, ngk) | 14.00 | 7.00 | 7.00 | 7.00 | 2.00 | 2.00 | 2.00 |
| Neisseria lactamica 020-06 (nla)a | 12.00 | 3.50 | 8.50 | 8.50 | 1.00 | 0.50 | 2.00 |
| Neisseria meningitidis (nme, nma, nmc) | 7.00 | 2.33 | 4.67 | 4.67 | 0.67 | 0.00 | 1.00 |
| Nocardia brasiliensis ATCC700358 (nbr) | 2.00 | 0.00 | 2.00 | 2.00 | 0.00 | 0.00 | 0.00 |
| Rickettsia peacockii Rustic (rpk)a | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Rickettsia prowazekii (rpr, rpo, rpw) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Rickettsia rickettsii (rrj, rra, rrc) | 3.00 | 2.00 | 1.00 | 1.00 | 1.00 | 0.00 | 1.00 |
| Salmonella bongori (sbg, sbz, sbv)a | 4.67 | 3.67 | 1.00 | 1.00 | 1.33 | 0.67 | 0.67 |
| Salmonella enterica subsp. enterica serovar typhi (sty, stt, sex) | 8.67 | 7.00 | 1.67 | 1.67 | 4.67 | 1.33 | 0.00 |
| Shigella flexneri (sfl, sfe, sfv, sfx) | 4.25 | 3.25 | 1.00 | 1.00 | 1.00 | 0.00 | 0.75 |
| Staphylococcus aureus (sau, saa, saj) | 3.00 | 3.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.00 |
| Streptococcus gordonii str. Challis substr. CH1 (sgo)a | 4.00 | 2.00 | 2.00 | 1.00 | 0.00 | 0.00 | 1.00 |
| Streptococcus pneumoniae (spn, spp, snt) | 5.00 | 4.33 | 0.67 | 0.67 | 0.33 | 0.00 | 2.00 |
| Streptococcus pyogenes (spy, spz, spk) | 3.33 | 2.33 | 1.00 | 1.00 | 0.00 | 0.00 | 1.00 |
| Treponema pallidum (tpa, tpp, tpw) | 1.00 | 1.00 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 |
| Treponema primitia ZAS-2 (tpi)a | 5.00 | 5.00 | 0.00 | 0.00 | 1.00 | 0.00 | 3.00 |
| Vibrio campbellii BAA-1116 (vha)a | 5.00 | 4.00 | 1.00 | 1.00 | 1.00 | 0.00 | 3.00 |
| Vibrio cholerae (vch, vcj, vco) | 5.33 | 4.33 | 1.00 | 1.00 | 1.00 | 0.00 | 2.33 |
| Yersinia pestis (ype, ypk, ypn) | 3.00 | 3.00 | 0.00 | 0.00 | 2.00 | 0.00 | 0.00 |
aNon-/less pathogenic bacteria
In following up with genome-reduced bacteria, we observed that only some subspecies of Mycoplasmagenitalium and Mycoplasma pneumoniae (mge and mpn) presented adenine MTases not associated with a restriction enzyme (K00571), and in the case of M. pneumoniae, these MTases have been reported as being non-essential in microtransposon experiments [17]. Additionally, only the adenine MTase associated with the type I restriction modification system (K03427) was present in Mycoplasma spp., and no other MTases associated with other restriction modification systems were observed. In the case of Rickettsia spp., only pathogenic Rickettsia rickettsii presented MTases; pathogenic R. prowazekii and non-pathogenic Rickettsia peacockii did not present a predicted MTase (Table 1). Together, these results could indicate that the reduced frequency of DNA MTases in some obligate intracellular bacteria with small genomes could be related to a lower dependency of DNA methylation either for the control of gene expression or restriction modification systems. This last possibility could be associated with a reduced likelihood of invasion by foreign DNA inside the host cell.
In considering the bacteria with the highest number of MTases, including H. pylori and Neisseria spp., some reports have mentioned an increased number of DNA MTases [15, 20] in the case of H. pylori, representing 4 % of its genome [15], and other authors have shown an association between some of these MTases (mod and cytosine MTases) with the expression of virulence-related genes [21, 22]. Although not all the predicted MTases in H. pylori are functional [15, 23], it is interesting to observe the epigenetic alterations reported in the mucosa of gastric cancers associated with H. pylori infection, including the hypermethylation of promoter sequences for the tumor suppressor genes WWOX [24], TFF2 and FOXD3 [20], suggesting the possibility that several functional DNA MTases could enter the host cell and methylate their recognition sequences in the host DNA, as previously mentioned [20]. Nevertheless, the increased expression of human DNA MTases DNMT1 and DNMT3 has also been shown in gastric cancer in comparison with controls, and even the expression of DNMT3a has been shown to be a poor prognosis indicator [25]. However, human DNA MTase overexpression has yielded controversial results when associated with H. pylori infection [24, 25]. In addition, H. pylori, N. meningitidis, N. gonorrhoeae and H. influenzae have shown the presence of phasevarions (a group of genes whose expression is controlled by a phase variable DNA MTase), which are associated with type III restriction modification systems [7, 21, 26] and are generally related to virulence and help bacteria to adapt better to their environment through a mechanism of reversible on/off gene expression switching; these mechanisms also help the bacteria to regulate their transformations by taking up foreign DNA [2]. We also observed that approximately half of the total number of DNA MTases corresponded to C5-cytosine MTases in Neisseria spp., which is a proportion that was only been observed in Bordetella petrii (according to our results), indicating a higher dependence of cytosine methylation in these bacteria.
With respect to our results, we were able to detect most, although not all, MTases reported in H. pylori [15, 23], and the mod gene associated with the type III restriction modification system could be detected in H. pylori and N. gonorrhoeae but not in N. meningitidis and H. influenzae. Nevertheless, these genes could be identified primarily as pseudo or hypothetical proteins in the NCBI/gene database for the analyzed genomes, and together with the previous results, they suggest the high specificity but relatively low sensitivity of the method implemented by KEGG to recognize homologs.
In relation to the other analyzed bacteria, we could corroborate the presence of Dam MTase in S. enterica serovar typhi, Yersinia pestis, V. cholerae, H. influenzae and Escherichia coli [2, 4, 27, 28] as well as its absence in Staphylococcus aureus [29]. An interesting observation is the strikingly high number of Dam MTases in S. enterica serovar typhi, which was the highest in the analyzed bacteria, suggesting a strong dependence of these enzymes on the regulation of gene expression, similar to that reported in S. enterica serovar typhimurium [2, 4] and S. enterica serovar enteritidis [30].
When we analyzed the descriptive and comparative results, we observed that most DNA MTases were adenine MTases with a significant increase over cytosine MTases, and around the half of these adenine MTases were not associated with a restriction modification system. These results are in accordance with the primary role of adenine MTases to regulate gene expression in bacteria [3]. We also observed that only adenine MTases were associated with a restriction modification system, and there was a higher number of DNA MTases associated with the type I restriction modification system compared to those associated with the type III restriction modification system, which were also more prevalent in all the analyzed bacteria, possibly indicating that these MTases are more required for restriction function, while MTases associated with the type III restriction modification system could be needed only by specific bacteria. This approach considers that some of these DNA MTases have evolved to exclusive modification MTases from a mutation in the restriction enzyme gene [31], and they help bacteria to better adapt as pathogens, as observed with the reported phasevarions associated with these enzymes. The presence of DNA MTases associated with the type II restriction modification system could not be measured because of their heterogeneous distribution among KO’s.
With respect to the correlations, we only found a significant correlation between the cytosine MTases and the genome size, however the correlation was low, indicating that the presence of DNA MTases is not importantly increased with the genome size and is more likely related to the specific epigenetic needs of each bacterium, which is probably associated with their lifestyle. However, it is important to mention that for the reduced genome bacteria, the number of most types of DNA MTases was significantly diminished, which could indicate a reduced dependency on these enzymes by intracellular obligate bacteria compared with the intracellular facultative or extracellular bacteria, although, as mentioned before, some of these enzymes could have been undetected.
In the comparison of related non-/less pathogenic bacteria, we observed that in some species, such as H. influenzae, H. pylori, E. coli O:157 and S. enterica serovar typhi, there was an evidently higher number of MTases compared with their non-/less pathogenic relatives; however, an opposite relation was also found for other bacteria, including Bordetella, Francisella, Neisseria and Treponema spp., which indicates that DNA MTases are highly variable in the pathogenic and the non-/less pathogenic species of the same genus depending on their particular evolution and lifestyle as well as that for some pathogenic species these MTases have evolved to regulate virulence genes, and they are therefore required for pathogenicity. This variability was also observed in the average of the S.D. for the global MTases per species, which was 1.16.
From all the MTases, only 7.29 % were encoded in a plasmid, indicating that most of these MTases are integrated into the genomic DNA, although many of these MTases could have been horizontally transmitted from other bacteria into plasmids or transposons and subsequently incorporated into the bacterial genome, as previously mentioned [15]. We observed that most part of DNA MTases (9/13) encoded in a plasmid belonged to the same KO (K07319), probably indicating that this KO is more likely to be horizontally transmitted.
In conclusion, we reported the general distribution of DNA MTases in human pathogenic bacteria and some non-/less pathogenic relatives, and we made important observations not previously mentioned, such as the low number of DNA MTases in genome-reduced bacteria, the significant increase in adenine MTases compared with cytosine MTases and the finding that only adenine MTases were associated with restriction modification systems. We also observed that DNA MTases related to restriction modification system type I were more prevalent than those associated with restriction modification system type III, and only a minority of MTases was encoded in a plasmid. In addition, we detected no correlation between the total number of DNA MTases and the genome size. Further reports designed to detect the presence of specific DNA MTases experimentally and to identify specific methylomes will clarify the distribution of DNA MTases in human pathogenic bacteria.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Organisms with a MamA homolog (XLSX 46 kb)
Consensus sequences of each KO analyzed. Footnote: The asterisks represent the catalytic amino acids (DOCX 91 kb)
Acknowledgments
We want to thank to DGAPA/UNAM for the postdoctoral scholarship granted to AJL Brambila-Tapia. Support from DGAPA-UNAM PAPIIT IN 107214 is also gratefully acknowledged.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Aniel Jessica Leticia Brambila-Tapia, Email: anieljessica@hotmail.com.
Katya Rodríguez-Vázquez, Email: katya.rodriguez@iimas.unam.mx.
References
- 1.Wion D, Casadesús J. N6-methyl-adenine: an epigenetic signal for DNA–protein interactions. Nat Rev Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar R, Rao DN. Role of DNA methyltransferases in epigenetic regulation in bacteria. Epigenetics: development and disease. Subcell Biochem. 2013;61:81–102. doi: 10.1007/978-94-007-4525-4_4. [DOI] [PubMed] [Google Scholar]
- 3.Casadesús J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Low DA, Weyand NJ, Mahan MJ. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun. 2001;69:7197–7204. doi: 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa K, Fukuda E, Kobayashi I. Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems. DNA Res. 2010;17:325–342. doi: 10.1093/dnares/dsq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seshasayee AS, Singh P, Krishna S. Context-dependent conservation of DNA methyltransferases in bacteria. Nucleic Acids Res. 2012;40:7066–7073. doi: 10.1093/nar/gks390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srikhanta YN, Dowideit SJ, Edwards JL, Falsetta ML, Wu HJ, Harrison OB, Fox KL, Seib KL, Maguire TL, Wang AH, Maiden MC, Grimmond SM, Apicella MA, Jennings MP. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog. 2009;5:e1000400. doi: 10.1371/journal.ppat.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breiling A, Lyko F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin. 2015;8:24. doi: 10.1186/s13072-015-0016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kekre A, Bhushan A, Kumar P, Kalia VC. Genome wide analysis for searching novel markers to rapidly identify clostridium strains. Indian J Microbiol. 2015;55:250–257. doi: 10.1007/s12088-015-0535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalia VC, Kumar P. Genome wide search for biomarkers to diagnose Yersinia infections. Indian J Microbiol. 2015;55:366–374. doi: 10.1007/s12088-015-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalia VC, Kumar R, Kumar P, Koul S. A gemone-wide profiling strategy as an aid for searching unique identification biomarkers for Streptococcus. Indian J Microbiol. 2015 doi: 10.1007/s12088-015-0561-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhushan A, Mukherjee T, Joshi J, Shankar P, Kalia VC. Insights into the origin of Clostridium botulinum strains: evolution of distinct restriction endonuclease sites in rrs (16S rRNA gene) Indian J Microbiol. 2015;55:140–150. doi: 10.1007/s12088-015-0514-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin LF, Posfai J, Roberts RJ, Kong H. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc Natl Acad Sci USA. 2001;98:2740–2745. doi: 10.1073/pnas.051612298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shell SS, Prestwich EG, Baek SH, Shah RR, Sassetti CM, Dedon PC, Fortune SM. DNA methylation impacts gene expression and ensures hypoxic survival of Mycobacterium tuberculosis. PLoS Pathog. 2013;9:e1003419. doi: 10.1371/journal.ppat.1003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lluch-Senar M, Luong K, Lloréns-Rico V, Delgado J, Fang G, Spittle K, Clark TA, Schadt E, Turner SW, Korlach J, Serrano L. Comprehensive methylome characterization of Mycoplasma genitalium and Mycoplasma pneumoniae at single-base resolution. PLoS Genet. 2013;9:e1003191. doi: 10.1371/journal.pgen.1003191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagar EA, Safarians S, Pang M. Analysis of genomic DNA from Chlamydia trachomatis for Dam and Dcm methylation. FEMS Microbiol Lett. 1992;77:161–168. doi: 10.1111/j.1574-6968.1992.tb05507.x. [DOI] [PubMed] [Google Scholar]
- 19.Hemavathy KC, Nagaraja V. DNA methylation in mycobacteria: absence of methylation at GATC (Dam) and CCA/TGG (Dcm) sequences. FEMS Immunol Med Microbiol. 1995;11:291–296. doi: 10.1111/j.1574-695X.1995.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 20.Sitaraman R. Helicobacter pylori DNA methyltransferases and the epigenetic field effect in cancerization. Front Microbiol. 2014;5:115. doi: 10.3389/fmicb.2014.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srikhanta YN, Gorrell RJ, Steen JA, Gawthorne JA, Kwok T, Grimmond SM, Robins-Browne RM, Jennings MP. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS One. 2011;6:e27569. doi: 10.1371/journal.pone.0027569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar R, Mukhopadhyay AK, Ghosh P, Rao DN. Comparative transcriptomics of H. pylori strains AM5, SS1 and their hpyAVIBM deletion mutants: possible roles of cytosine methylation. PLoS One. 2012;7:e42303. doi: 10.1371/journal.pone.0042303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krebes J, Morgan RD, Bunk B, Spröer C, Luong K, Parusel L, Anton BP, König C, Josenhans C, Overmann J, Roberts RJ, Korlach J, Suerbaum S. The complex methylome of the human gastric pathogen Helicobacter pylori. Nucleic Acids Res. 2014;42:2415–2432. doi: 10.1093/nar/gkt1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan J, Zhang M, Zhang J, Chen X, Zhang X. Helicobacter pylori infection promotes methylation of WWOX gene in human gastric cancer. Biochem Biophys Res Commun. 2011;408:99–102. doi: 10.1016/j.bbrc.2011.03.127. [DOI] [PubMed] [Google Scholar]
- 25.Cao XY, Ma HX, Shang YH, Jin MS, Kong F, Jia ZF, Cao DH, Wang YP, Suo J, Jiang J. DNA methyltransferase3a expression is an independent poor prognostic indicator in gastric cancer. World J Gastroenterol. 2014;20:8201–8208. doi: 10.3748/wjg.v20.i25.8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox KL, Dowideit SJ, Erwin AL, Srikhanta YN, Smith AL, Jennings MP. Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res. 2007;35:5242–5252. doi: 10.1093/nar/gkm571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gómez-Eichelmann MC. Deoxyribonucleic acid adenine and cytosine methylation in Salmonella typhimurium and Salmonella typhi. J Bacteriol. 1979;140:574–579. doi: 10.1128/jb.140.2.574-579.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson VL, Oyston PC, Titball RW. A dam mutant of Yersinia pestis is attenuated and induces protection against plague. FEMS Microbiol Lett. 2005;252:251–256. doi: 10.1016/j.femsle.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Dreiseikelmann B, Wackernagel W. Absence in Bacillus subtilis and Staphylococcus aureus of the sequence-specific deoxyribonucleic acid methylation that is conferred in Escherichia coli K-12 by the dam and dcm enzymes. J Bacteriol. 1981;147:259–261. doi: 10.1128/jb.147.1.259-261.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarnacki SH, Castañeda MDELR, Noto Llana M, Giacomodonato MN, Valvano MÁ, Cerquetti MC. Dam methylation participates in the regulation of PmrA/PmrB and RcsC/RcsD/RcsB two component regulatory systems in Salmonella enterica serovar Enteritidis. PLoS One. 2013;8:e56474. doi: 10.1371/journal.pone.0056474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srikhanta YN, Fox KL, Jennings MP. The phasevarion: phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat Rev Microbiol. 2010;8:196–206. doi: 10.1038/nrmicro2283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Organisms with a MamA homolog (XLSX 46 kb)
Consensus sequences of each KO analyzed. Footnote: The asterisks represent the catalytic amino acids (DOCX 91 kb)