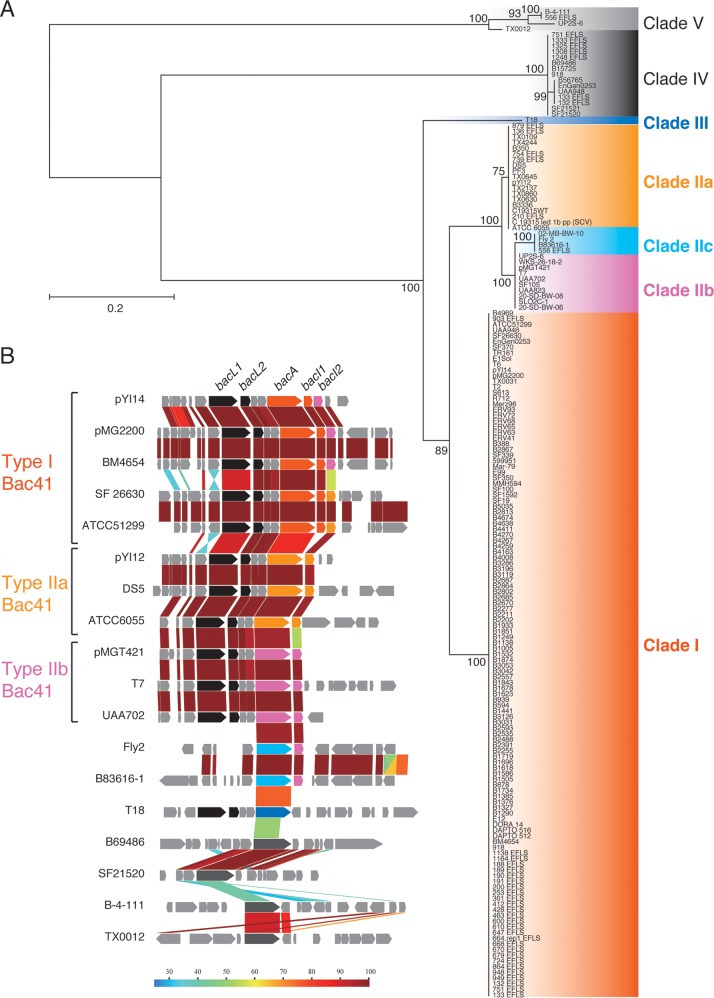
FIG 2.
Diversity of BacA proteins in E. faecalis strains. (A) The phylogenic tree of BacA homologues of E. faecalis strains was constructed using the JTT model in MEGA7 based on the amino acid sequence alignment generated by ClustalW version 2.0. The tree with the highest log likelihood is shown. The percentage of trees in which the associated taxa clustered together is shown next to each branch. The initial tree(s) for the heuristic search was obtained automatically by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model and then selecting the topology with the superior log-likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 174 amino acid sequences. All positions containing gaps and missing data were eliminated. The names of the source strains are shown. (B) Genetic structure alignment for the flanking regions of the respective BacA homologues. Color scale represents amino acid sequence similarity (%). Conserved CDSs, including bacL1 and bacL2, are represented in black. Specific CDSs including bacA, bacI1, and bacI2 are represented as follows: type I, orange; type IIa, yellow; type IIb, pink; type IIc, cyan; type III, blue; types IV and V, gray.