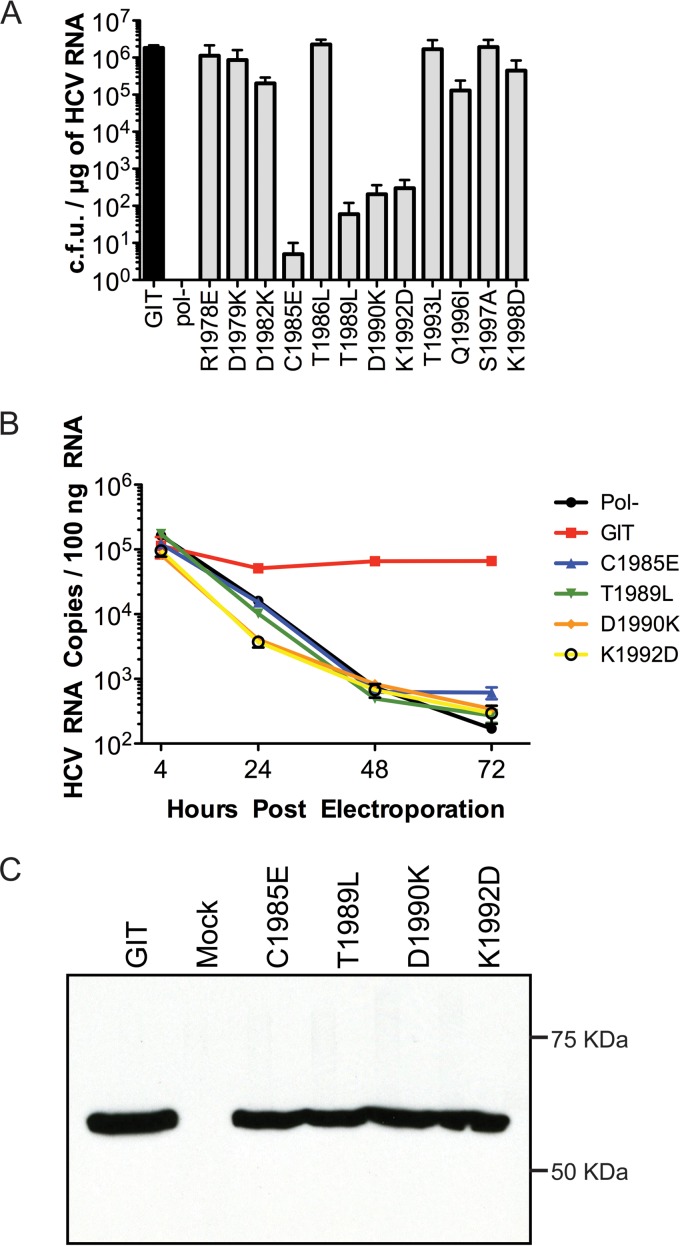
FIG 2.
Characterization of NS5A anchor helix single mutants. (A) RNA replication efficiencies of the replicons bearing mutations in the NS5A anchoring helix region. The bar diagram represents the transduction efficiencies of replicon RNAs in CFU per microgram of HCV RNA. pol(−) indicates a Con1/SG-neo replicon RNA bearing a nonfunctional RNA-dependent RNA polymerase sequence that served as a negative control, while the transduction efficiency of the parental Con1/SG-neo GIT replicon (labeled GIT) served as a positive control for replication experiments. The assay was performed in triplicates, and each bar from the graph represents the average number of colonies (with standard deviations) obtained from three experimental replicates for each experimental mutant and control replicons. (B) Assessment of transient RNA replication of selected mutants (mutants identified as lethal or near lethal by a replication assay [C1985E, T1989L, D1990K, and K1992D]) of HCV RNA by real-time RT-PCR. The numbers of HCV RNA copies present in the case of each experimental mutant per 100 ng of total RNA extracted at 4 h, 24 h, 48 h, and 72 h postelectroporation are plotted in the graph. Each sample was assayed by RT-PCR in triplicates. (C) Following transient expression, NS5A processing for the lethal or near-lethal mutants was analyzed by Western blotting with an anti-NS5A monoclonal antibody (9E10).