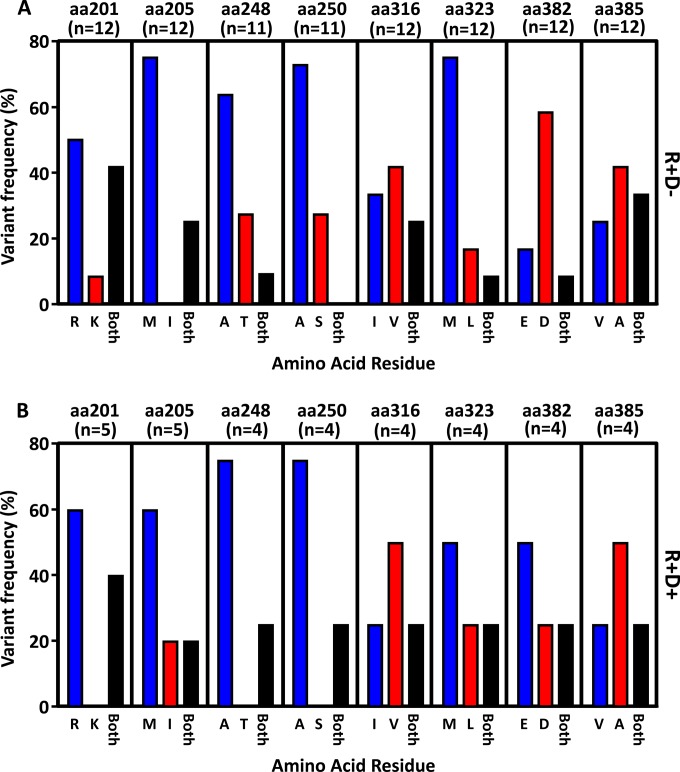
FIG 1.
Pyrosequencing analysis of the IE-1 sequence variants in HSCT recipients. DNA was extracted from plasma samples of 17 HSCT recipients during CMV reactivation. Following DNA PCR amplification, pyrosequencing analysis of the panel of SNPs was performed as outlined in Materials and Methods. The nucleotide data were extrapolated to determine the proportion of each amino acid residue for the 8 positions tested. (A) Data represent the proportion of R+/D− recipient samples encoding either a dominant single amino acid residue at each position or both amino acid residues at each position. (B) Data represent the proportion of R+/D+ recipient samples encoding either a dominant single amino acid residue at each position or both amino acid residues at each position.