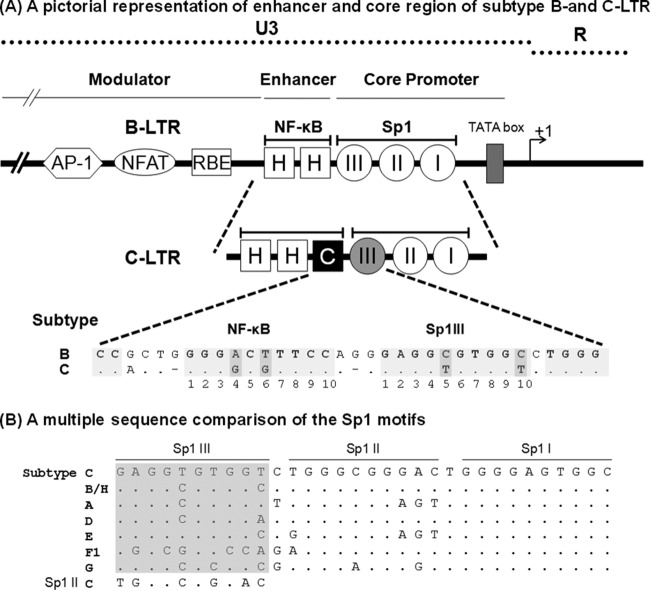
FIG 1.
(A) Schematic diagram comparing the TFBS of the enhancer and core promoter elements of B- and C-LTRs. An outline of the prototype B LTR with some of the basic transcription factor binding sites is shown at the top (not to scale). The enhancer region of the B-LTR contains two κB motifs (HH), in contrast to the C-LTR, which contains three κB elements (HHC). The additional NF-κB motif is genetically distinct and is referred to here as the C-κB site (filled square box). Note that the enhancer-proximal Sp1III site is also characterized by subtype-specific genetic variations, as shown at the bottom. The central TFBS sequences are shaded, and the genetic variations between the subtypes are highlighted by the darker shading. Additionally, the spacer length between the two TFBS is comprised of only two bases in subtype C, unlike the three bases in subtype B. (B) Multiple-sequence comparison of the Sp1 motifs. The nucleotide sequences of Sp1III, Sp1II, and Sp1I motifs representing the consensus sequences of the major HIV-1 genetic subtypes are aligned. Dots in the alignment represent sequence identity. The sequence alignment of the Sp1III motif is highlighted in gray. The Sp1II sequence of subtype C is also included in the alignment of Sp1III sequences for comparison.