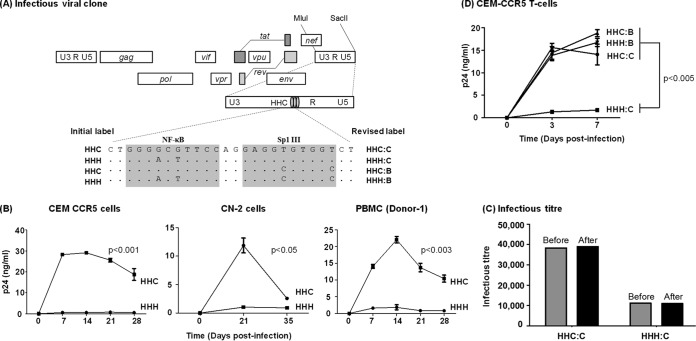
FIG 9.
Replication profiles of the NF-κB and Sp1III chimera viral strains in target cells. (A) The schematic representation of a panel of NF-κB site variant infectious viral vectors constructed using Indie C1. The centrally positioned NF-κB and Sp1III motifs in the 3′ LTR are highlighted by shading, and the genetic variations engineered into these elements are shown. Of the four viral clones, the top two (HHC and HHH) were constructed for the early rounds of the assays, and two additional clones were constructed subsequently. The initial labeling of the first two vectors is shown on the left side and the revised labeling of all four viral strains on the right side. The letters B and C following the colon in the label represent the subtype identity of the Sp1III motif. In all four variant promoters, the other two Sp1 motifs (II and I) are intact but are not shown. Dots in the sequence alignment represent sequence homology. (B) CEM-CCR5 T cells, CN-2 cells, herpesvirus saimiri strain-transformed CN-2 cells, or CD8+ cell-depleted and mitogen-activated PBMC from a healthy donor were infected with 500 TCID50 of Indie-HHC or Indie-HHH virus. The secretion of p24 into the culture medium was monitored at regular intervals for several weeks. (C) A titer comparison of the viral stocks before and after the assay. The TCID50 titers of HHC and HHH viral strains were determined in TZM-bl cells using β-galactosidase staining before and 4 weeks after the target cell infection to determine the replication profile of the viruses performed as described for panel A. Each bar represents the means from triplicate wells. The data are representative of two independent experiments. (D) Replication profile of all four promoter variant viral strains in CEM-CCR5 cells. The data are presented as means of triplicate wells ± SD and are representative of at least three independent experiments. A two-tailed unpaired t test was used for statistical analysis.