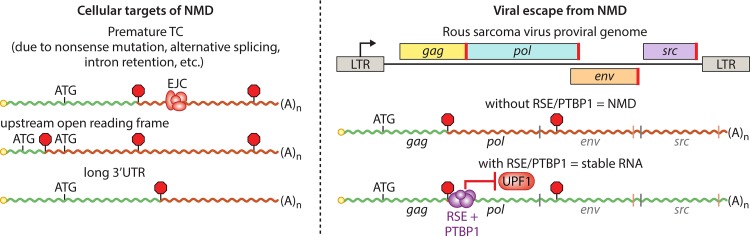
FIG 2.
Determinants of cellular and viral RNA susceptibility to NMD. (Left) RNA features leading to induction of NMD. In each case, translation termination occurs at sites distant from the 3′ end of the transcript. Decay is accelerated if termination takes place upstream of an EJC. (Right) RSV full-length mRNAs use the RSE to recruit host PTBP1 protein to the vicinity of the gag stop codon, preventing UPF1 association and inhibiting NMD. If the RSE is deleted or unable to bind PTBP1, RSV mRNAs are efficiently degraded by NMD. Positions of TCs typically used in full-length RNAs are indicated by stop signs, and positions of start and stop codons of additional ORFs are shown with light gray and red vertical lines, respectively. Red segments of RNAs indicate regions that may be recognized by UPF1 as aberrantly long 3′UTRs. LTR, long terminal repeat.