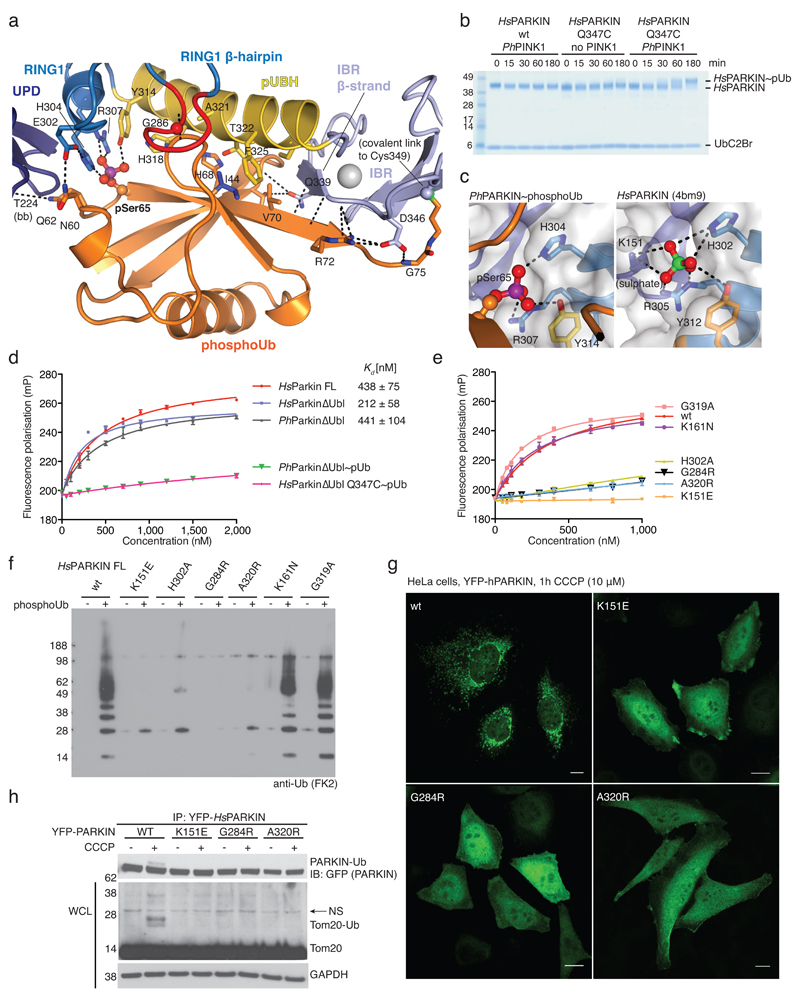
Figure 2. PhosphoUb binding to PARKIN.
(a) PhosphoUb binding site on PhPARKIN as in Fig. 1c. Dotted lines indicate hydrogen bonds. The RING1 β-hairpin that harbours patient mutations, is highlighted in red. bb, backbone contacts. (b) PhosphoUb suicide probe reactions as in Fig. 1b with UbC2Br and GST-PhPINK1. The experiment was performed three times with consistent results. (c) (Left) Occupied Ser65 phosphate pocket in PhPARKIN, (right) identical pocket in HsPARKIN occupied by a sulphate ion in PDB 4bm9 14. (d) Fluorescence polarisation experiments characterizing the binding of FlAsH-tagged phosphoUb to PARKIN variants. Measurements were performed in triplicate and error bars represent standard deviation from the mean. mP, milli-polarization unit. (e) Binding assays as in d with full-length HsPARKIN and mutants in the phosphoUb binding site. Binding curves were compiled from experiments shown in Extended Data Fig. 3d. Measurements were performed in triplicate and error bars represent standard deviation from the mean. (f) Activity assays of full-length HsPARKIN variants with and without phosphoUb. After 2 h, reactions were resolved by SDS-PAGE and polyubiquitin visualised by anti-polyubiquitin Western blotting (FK2, Millipore). PARKIN protein normalization is shown in Extended Data Fig. 3e. The experiment was performed three times with consistent results. (g) YFP-HsPARKIN wt or mutants were transfected into HeLa cells, treated with CCCP (10 µM) for 1 h and visualised by immunofluorescence. See Extended Data Fig. 4 for controls and quantification. Scale bar, 10 μm. (h) HeLa cell lysates expressing YFP-HsPARKIN wt or mutants were western blotted for PARKIN (after immunoprecipitation (IP)) and Tom20 (in whole-cell lysate (WCL)). NS, non-specific band. The experiment was performed at least twice as biological replicate for every mutant with consistent results. See Extended Data Fig. 4d and Supplementary Information.