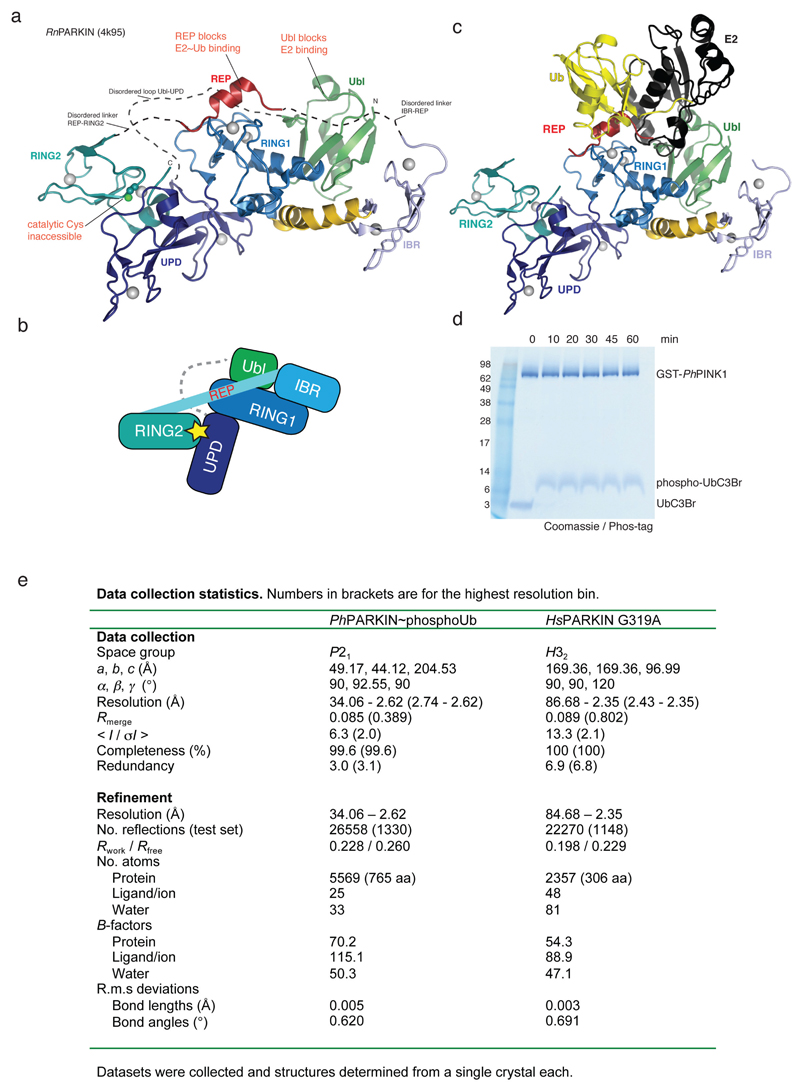
Extended Data Figure 1. Autoinhibited PARKIN and phosphoUb probes.
(a) Structure of rat PARKIN (RnPARKIN, pdb-id 4k95, 16, chain A is used for all representations of RnPARKIN) with Ubl domain coloured in green, the Unique PARKIN domain (UPD) in dark blue, RING1 in blue, IBR in lightblue, REP in red, and RING2 in cyan. Zinc atoms are shown as grey spheres. The catalytic Cys431 is shown in ball-and-stick representation. Disordered linkers are indicated as dotted lines. The three mechanisms of PARKIN inhibition are pointed out in red. (b) Schematic diagram of the closed, autoinhibited conformation of full-length PARKIN. (c) Superposition of the E2~Ub complex from the BIRC7 structure (pdb-id 4auq, 35), superposed via its RING domain onto RING1 of full-length RnPARKIN (pdb-id 4k95, 16) to indicate the position of the E2~Ub on PARKIN RING1. Assuming that E2~Ub adopts a canonical conformation on RING1, the E2 would clash with the Ubl domain and partially with the REP, while the E2-linked Ub would clash with the REP. Hence, Ubl and REP have to be released to enable PARKIN E2~Ub binding at RING1. (d) Time course analysis of an exemplary reaction of UbC3Br (0.2 mg/ml) phosphorylated by GST-PhPINK1 (5 µM) as described previously for ubiquitin8. Phosphorylation of proteins was monitored by a band shift on Coomassie-stained Phos-Tag gels. The experiment was performed two times with consistent result. (e) Data collection and refinement statistics.