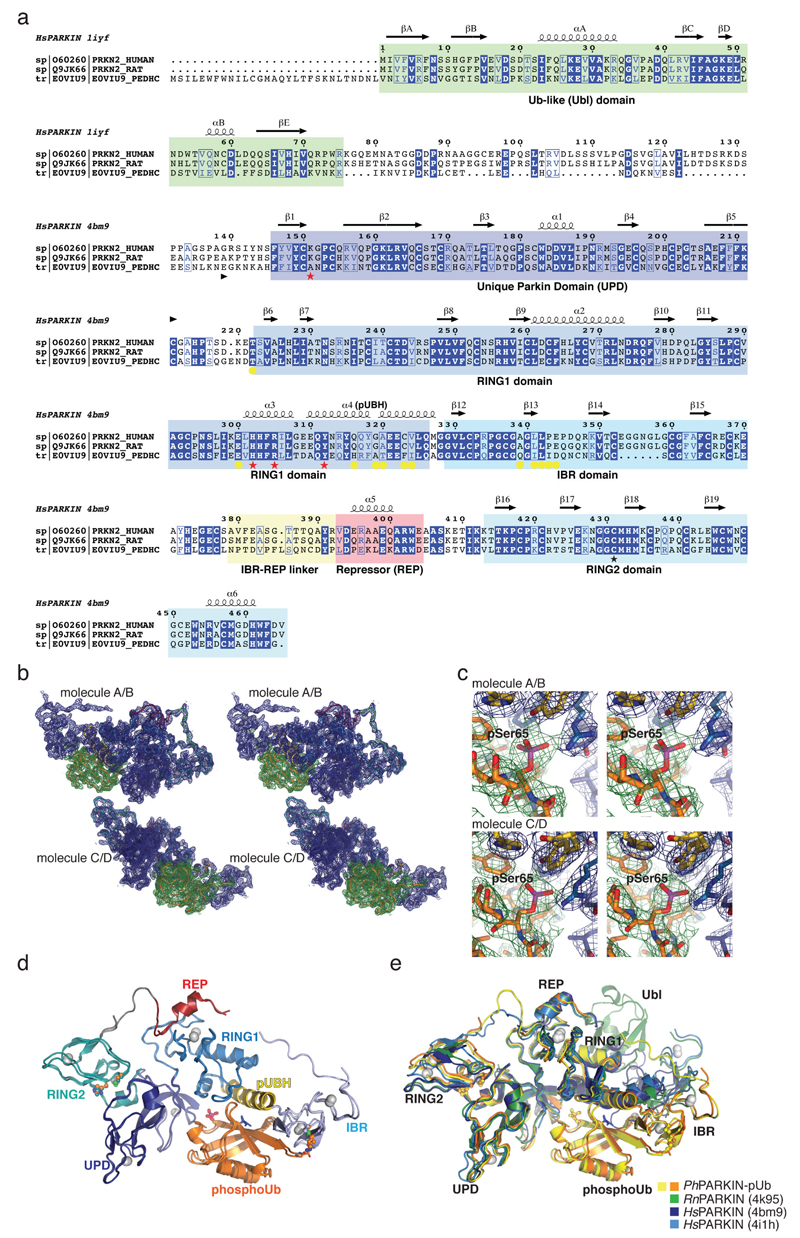
Extended Data Figure 2. PhPARKIN similarity with human and rat PARKIN and map quality.
(a) Structure-based sequence alignment of PARKIN from human (top), rat (middle) and Pediculus humanus corporis (PhPARKIN, bottom). The domains are indicated in boxes coloured according to structural figures. HsPARKIN and PhPARKIN are 45% identical within their crystallised constructs. Secondary structure elements are shown for HsPARKIN Ubl domain (pdb-id 1iyf, 36) and for HsPARKIN core domain (4bm9, 14). Red stars denote the phospho-Ser65 Ub binding pocket, and yellow spheres the residues contacting phosphoUb (see Fig. 2). A black star denotes the catalytic Cys in RING2. (b) Stereo representation of the asymmetric unit of PhPARKIN~pUb crystals, showing 2|Fo|-|Fc| electron density at 1σ, in blue for PhPARKIN and in green for phosphoUb. (c) Electron density detail, shown as in b, zooming in on phosphoUb phospho-Ser65, in stereo representation (d) Superposition of the two PhPARKIN~pUb complexes in the asymmetric unit, coloured as in Fig. 1. The RMSD is 0.76 Å. Electron density is missing for parts of the flexible linker between IBR and RING2. (e) Superposition of available PARKIN structures (4bm9 14; 4i1h 15; 4k95 16 and two PhPARKIN~pUb complexes) in different colours, showing similar domain positions with respect to each other, with exception of the IBR domain. Only the structure of full-length RnPARKIN16 contains the Ubl domain.