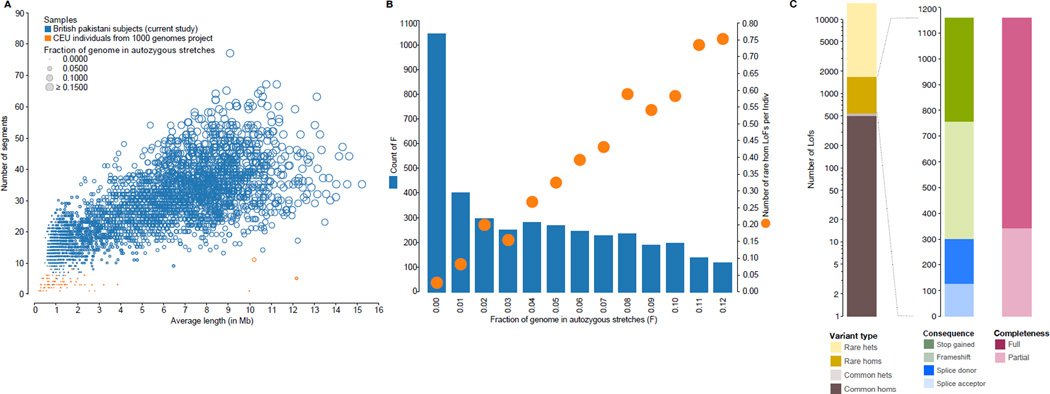
Fig. 1. Discovery and annotation of rhLOF variants.
(A) Autozygous segment numbers and length for Pakistani heritage subjects in the UK, and 1000 Genomes project European (CEPH Utah residents with ancestry from northern and western Europe; CEU) individuals. (B) Autozygosity and rhLOF in 3,222 individuals. Count of number of individuals (left Y axis, blue columns) binned by fraction of autozygous genome (X axis, showing values from 0.00 to 0.12), with mean number of rhLOF genotypes per individual (right Y axis, orange circles). (C) Distribution of LOF variants by allele frequency, heterozygous or homozygous genotype, predicted protein consequence, and whether predicted for a full or partial set of GENCODE Basic transcripts for the gene.