Figure 2.
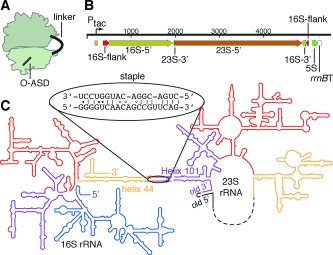
Ribosome subunit stapling. A) Illustration of the stapled ribosome. B) 1‐D representation of the designed rDNA operon. The 16S gene is separated into two halves (16S‐5′ and 16S‐3′) by the insertion of 23S rDNA. The 23S gene is circularly permuted such that a 3′‐terminal segment (23S‐3′) precedes its 5′‐terminal segment (23S‐5′). This transcript (ca. 4500 nt) is flanked by the native 16S processing sites (flanks). The 23S processing sites (which would normally liberate the 23S rRNA into a separate RNA molecule) have been deleted. C) 2‐D representation of the stapled ribosome, illustrating the linker sequence used to staple helix 44 (h44) of 16S rRNA to Helix 101 (H101) of 23S rRNA into a single rRNA molecule.
