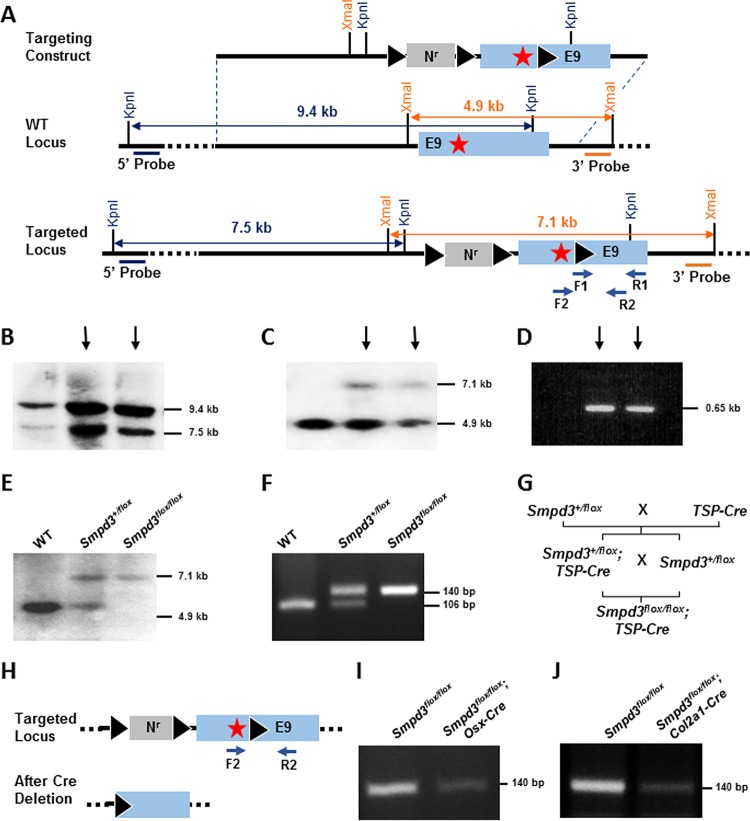
FIG 7.
Tissue-specific deletion of Smpd3 gene. (A) Schematic representation of the targeting construct and the targeted Smpd3 locus. A neomycin resistance cassette (Nr) flanked by two loxP sites and a separate third loxP site were inserted into intron 8 and exon 9 downstream of the stop codon (star), respectively. 5′ and 3′ probes are indicated, and the primers (F1, R1, F2, and R2) used for PCR analyses are shown by arrows. KpnI and XmaI restriction sites were used for the Southern blot screening by the 5′ and 3′ probes, respectively. (B) ES cell clones with homologous recombination at the Smpd3 locus were identified by Southern blotting using the 5′ probe. The 9.4- and 7.5-kb bands represent the KpnI fragments generated from the WT and the targeted locus, respectively. (C) The screening of the targeted clones using the 3′ probe shows 4.9-kb and 7.1-kb bands denoting the XmaI fragments generated from the WT and the targeted locus, respectively. (D) PCR amplification of ES cell DNA using F1 and R1 primers showing amplification of a 0.65-kb fragment from the targeted clones containing the third loxP site. (B to D) Arrows indicate the clones used for microinjection. (E) Southern blot analysis of WT, heterozygote (Smpd3+/flox), and homozygote (Smpd3flox/flox) mice in the same litter using the 3′ probe. (F) Genotyping PCR using F2 and R2 primers to detect the targeted Smpd3 floxed allele (140 bp) and the WT allele (106 bp). (G) Breeding scheme showing the generation of Smpd3flox/flox; Osx-Cre and Smpd3flox/flox; Col2a1-Cre mice. TSP-Cre, tissue-specific Cre. (H) Schematic representation of the targeted locus before and after deletion of the floxed sequence in the presence of Cre recombinase. Arrows, the primers used for PCR analysis. (I and J) PCR to detect the deletion of the floxed Smpd3 sequence shown by a decrease of band intensity in the calvaria of Smpd3flox/flox; Osx-Cre mice and in the cartilage of Smpd3flox/flox; Col2a1-Cre mice.