Abstract
The SWI/SNF and RSC family of ATP-dependent chromatin remodelers disassembles nucleosomes by moving nucleosomes into the vicinity of adjoining nucleosomes. We found that the histone chaperone Nap1 efficiently promotes disassembly of adjacent nucleosomes with which RSC collides and not the disassembly of nucleosomes mobilized by RSC. Nap1 is specific to RSC, as it does not target SWI/SNF, its paralog in Saccharomyces cerevisiae. Extensive mutational analysis of Nap1 has revealed that Nap1 affinity for histones H2A-H2B and H3-H4 and its ability to displace histones from DNA are required for Nap1 to enhance RSC-mediated disassembly. Other histone chaperones, such as Vps75, that also bind histones are not able to enhance RSC-mediated disassembly. Our study suggests a mechanism by which Nap1 is recruited to actively transcribed regions and assists in the passage of the transcription complex through chromatin, and it provides a novel mechanism for the coordinated action of RSC and Nap1.
INTRODUCTION
Nucleosomes, the building blocks of chromatin, consist of a central octameric core formed from two copies of each of the core histone proteins (H2A, H2B, H3, and H4) wrapped nearly twice with approximately 147 bp of DNA. The core of one nucleosome is separated from the adjacent nucleosome via a segment of linker DNA. Nucleosomes are stable in vitro but are rapidly exchanged in vivo independent of DNA replication. Nucleosome exchange is mediated by chromatin remodelers and histone chaperones (1–3). Several histone chaperones contribute to nucleosome assembly and disassembly; however, it is not clear how nucleosomes are turned over and what determines the balance between nucleosome assembly and disassembly. ATP-dependent chromatin remodelers are also involved in the assembly and disassembly of nucleosomes. The chromatin-remodeling complex RSF assembles nucleosomes without additional factors, whereas the chromatin-remodeling complex ACF organizes prenucleosomes that have been deposited onto DNA by the histone chaperone Nap1 (4–7). In contrast, the SWI/SNF family of chromatin remodelers disassembles nucleosomes from chromatin (8).
We have previously shown that efficient nucleosome disassembly by the RSC and SWI/SNF complexes requires a minimum of two contiguous nucleosomes but that only one of these two nucleosomes is displaced (9, 10). SWI/SNF and RSC mobilize one nucleosome, with the adjacent nucleosome being displaced when the mobilized nucleosome moves into its vicinity. Although several studies have uncovered potential mechanistic synergy between histone chaperones and ATP-dependent chromatin remodelers in destabilizing and disassembling chromatin (11–14), the details of these mechanisms are unclear.
Nap1 was first identified as a factor from mammalian HeLa cell extract that assists in nucleosome assembly in vitro (15). Nap1 is highly conserved among eukaryotes, and Nap1 knockouts result in embryonic lethality in Drosophila and mouse (16, 17). In Saccharomyces cerevisiae, loss of Nap1 alters the expression of 10% of its genes (18), and Nap1 is implicated in cell cycle and transcription regulation, exchange of histone variants, chromatin assembly, and promotion of nucleosome sliding. Nap1 is thought to be primarily an H2A-H2B chaperone but can also bind H3-H4 heterotetramers and the linker histone H1 (19–23).
The crystal structure of Nap1 shows a signature fold composed of α-helices and β-strands with a long α2 helix mediating homodimerization (24). The dimer has an overall ellipsoidal shape with a negatively charged surface responsible for histone binding (25). Nap1 competes for H2A-H2B binding to DNA in vitro, yet Nap1 cannot recognize the surface of H2A-H2B in canonical nucleosomes because of shielding by DNA. Consequently, it is not known how Nap1 gains access to H2A-H2B in order to disassemble nucleosomes.
Nap1 appears to work in conjunction with ATP-dependent chromatin remodelers, such as Chd1 and RSC, in regulating chromatin organization. In Schizosaccharomyces pombe cells, Nap1 works cooperatively with Chd1 to disassemble nucleosomes in promoter regions (26). Nap1, when in high concentration, has also been suggested to act in concert with RSC to disassemble mononucleosomes in vitro (13). Other data suggest that Nap1 evicts H2A-H2B from mononucleosomes when remodeled by RSC (14).
We find that Nap1, at relatively low concentrations, enhances RSC-mediated nucleosome disassembly of dinucleosomes, but not mononucleosomes, by a mechanism unrelated to an increased rate of ATP hydrolysis or nucleosome movement by RSC. Instead, we find that Nap1 influences later steps in the movement of the adjacent nucleosome. The region of Nap1 critical for this activity is the region previously shown to bind to H2A-H2B and/or to facilitate its competition from DNA The competition between histone binding to Nap1 or DNA appears to be critical for Nap1 to enhance RSC-mediated nucleosome disassembly. Finally, the coordinated action between Nap1 and RSC is specific, as this function cannot be replaced by either the Nap1 homolog Vps75 or the chromatin remodeler SWI/SNF.
MATERIALS AND METHODS
Purification of RSC-FLAG, His-Nap1, other chaperones, yeast histones, and octamer.
The RSC remodeling complex was purified from S. cerevisiae (YNC001) using C-terminally FLAG-tagged Rsc2 and an anti-FLAG immunoaffinity purification procedure (27). Briefly, 60 liters of yeast cells were grown to an optical density at 600 nm (OD600) of ∼5 and harvested at 4,000 rpm for 10 min. The cell pellet was washed with water and extraction buffer and frozen with liquid nitrogen into spaghetti. The yeast spaghetti was ground to fine powder using a Spex grinding mill, buffer was added and stirred, and the supernatant was collected after centrifugation at 100,000 × g. An extract was incubated with anti-FLAG M2–agarose beads (Sigma-Aldrich) overnight at 4°C with mixing. After several rounds of washing, the protein was eluted in buffer containing 1 mg/ml FLAG peptide. The expression plasmid for N-terminally 6×His-tagged yeast Nap1 was kindly provided by Toshio Tsukiyama. Nap1 was overexpressed in Escherichia coli by induction at an OD600 of ∼0.2 with IPTG (isopropyl-β-d-thiogalactopyranoside). Purification was performed using Ni-nitrilotriacetic acid (NTA) resin and 250 mM imidazole for elution. The protein fractions were dialyzed twice for 2 h each time before changing the buffer. Fractions were then dialyzed overnight against the same elution buffer containing 10% glycerol rather than imidazole. Nap1 mutants were in a cysteine mutant background (C200A, C249A, and C272A) and were purified as described previously (25). Vps75 was purified as described previously (28). Purified recombinant MCM2 protein was kindly provided by Dinshaw Patel's laboratory at Memorial Sloan Kettering Cancer Center. Recombinant yeast histones H2A, H2B, H3, and H4 were purified as described previously (29) and refolded into octamers by dialysis prior to purification by gel filtration using Superdex 200 (GE; prep grade). For site-directed mapping experiments, H2B with cysteine at residue 53 was generated by site-directed mutagenesis. H2B histones containing cysteine were combined with other wild-type (WT) histones. Importantly, the native cysteine residue H3-110 had previously been mutated to alanine, and thus, H2B S53C was the sole cysteine residue present in all four histones.
Nucleosome reconstitution.
Mononucleosomes (29N59) were assembled with 235-bp DNA containing the 601 positioning sequence flanked by 29 and 59 bp of extranucleosomal DNA. Dinucleosomes [40-N(601)-31-N(603)-6] were assembled using a 371-bp DNA fragment containing the 601 and 603 positioning sequences separated by a 31-bp intervening sequence, 40 bp of flanking DNA at the 601 end, and 6 bp of flanking DNA at the 603 end. Within the 40 bp of flanking DNA, a single Gal4 site was positioned 23 bp from the nucleosome. DNA for mono- and dinucleosomes was prepared by PCR from p159-1Gal4-27 and p159-1Gal4-27-601-603 plasmid DNA templates, respectively, followed by phenol-chloroform extraction and ethanol precipitation. The DNA was further purified by ultracentrifugation with an Amicon 30K centrifugal filter unit (27). Nucleosomes were assembled by the rapid salt dilution method (2 M to 190 mM NaCl) at 25°C, using WT yeast histone octamers, and analyzed on 4% (35.4:1 acrylamide to bisacrylamide) native gels (29). Dinucleosomes for mapping histone-DNA contacts were assembled as described previously using recombinant Xenopus laevis histones (27).
Binding assay.
A 100-bp DNA fragment was generated by PCR amplification from plasmid p199 and purified with a PCR cleanup kit (VWR) before being used for binding and ATPase assays. WT yeast dinucleosomes [40-N(601)-31-N(603)-6] or 20 nM 100-bp DNA was used as the substrate. RSC was titrated from 5 to 40 nM to define saturated binding conditions for remodeling and ATP hydrolysis experiments with Nap1. For binding, Nap1 was titrated from 30 nM to 9 μM. DNA or dinucleosomes, RSC, and varying amounts of Nap1 were added together, incubated at 30°C for 15 min, loaded on 4% native PAGE gels (79:1 acrylamide to bisacrylamide) that ran at 200 V in 0.5× Tris-borate-EDTA (TBE). Reaction mixtures contained 20 mM HEPES-NaOH (pH 7.8), 3 mM MgCl2, 6% (vol/vol) glycerol, 70 mM NaCl, and 0.1 mg/ml bovine serum albumin (BSA). The mixture for the reciprocal experiment, using a fixed amount of Nap1 and varying amounts of RSC, contained 20 nM 100-bp DNA, 300 nM Nap1, with 5 to 40 nM RSC added. Gel images were quantified using Optiquant software, and the percentage of DNA or dinucleosomes bound was plotted against either the fold molar excess of Nap1 or the concentration of RSC. Binding curves were generated in GraphPad (Prism) and fitted to a single-site binding model.
Remodeling time course.
Reaction mixtures contained 20 nM dinucleosomes, 20 nM RSC, 300 nM Nap1 (WT or mutant), and 55 μM ATP. At each time point, 1 μl of a stop mixture containing 5 μg sheared salmon sperm DNA and 5 mM EDTA was added to 15 μl of the reaction mixture. For consistency, after stopping the reaction, 300 nM Nap1 was added to the reaction mixtures lacking Nap1. Samples were analyzed on a 4% native gel (35.4:1 acrylamide to bisacrylamide). Disappearance of the original nucleosomal band or appearance of remodeled species II over time was plotted using GraphPad (Prism). The data were fitted to a one-phase exponential association using GraphPad (Prism).
ATPase assay.
The rate of ATP hydrolysis was determined under saturating conditions for either RSC or DNA. ATPase assays with saturating amounts of DNA contained 4 nM RSC and 20 nM 100-bp DNA; reactions with saturating amounts of enzyme contained 40 nM RSC and 20 nM 100-bp DNA. Nap1 was added at molar excesses of 5- and 15-fold with respect to DNA or RSC. For the dinucleosome-stimulated ATPase assay, reaction mixtures contained 20 nM dinucleosomes, 20 nM RSC, 55 μM ATP, and varying amounts of Nap1. The background signal was measured using samples lacking the remodeler. Additionally, any intrinsic background hydrolysis observed with remodeler in the absence of nucleosomes was subtracted from the signal. [γ-32P]ATP was added to the reaction mixtures, and hydrolysis was stopped at regular time intervals with SDS (1.5%) and EDTA (50 mM). Inorganic phosphate and ATP were separated by thin-layer chromatography on polyethyleneimine cellulose plates (J. T. Baker, Germany) developed with 0.5 M LiCl and 0.5 M formic acid.
Site-directed mapping.
Site-directed histone-DNA cross-linking was performed as described previously (30). Briefly, histone octamers containing cysteine at residue 53 of H2B were reconstituted into dinucleosomes and conjugated to p-azidophenacyl bromide (APB) from Sigma. RSC in the absence and presence of Nap1 was incubated with APB-modified dinucleosomes under full binding conditions at 30°C for 10 min. For remodeling experiments, samples included 10 μM ATP, and the remodeling reactions were stopped with excess EDTA (15 mM). Samples were UV cross-linked at 312 nm for 3 min before adding SDS to a final concentration of 1% and incubating at 37°C for 15 min. DNA cross-linked to histones was enriched through phenol-chloroform extraction and ethanol precipitation prior to resuspension in 1 M pyrrolidine (Sigma). Samples were incubated at 90°C for 15 min, and the solvent was removed with a Centrivap concentrator (Labconco). The 5′ end-labeled DNA was resuspended in 25 μl ultrapure water and dried in a Centrivap twice before resuspension in 10 μl 95% formamide, 0.0625% bromophenol blue and xylene cyanol. DNA samples were analyzed on a 6.5% denaturing polyacrylamide gel, along with a sequence ladder of the same DNA; visualized by phosphorimaging; and quantified with ImageQuant (version 5.2; Packard Instrument Company). Lane intensities were normalized for loading bias using Microsoft Excel. Band intensities were plotted as a function of time and fitted into a one-phase decay or one-phase association equation using GraphPad (Prism; version 6.0b). Overlays for different time points were made using the ggplot2 package in R.
RESULTS
Nap1 selectively enhances RSC-mediated disassembly of dinucleosomes but not mononucleosomes.
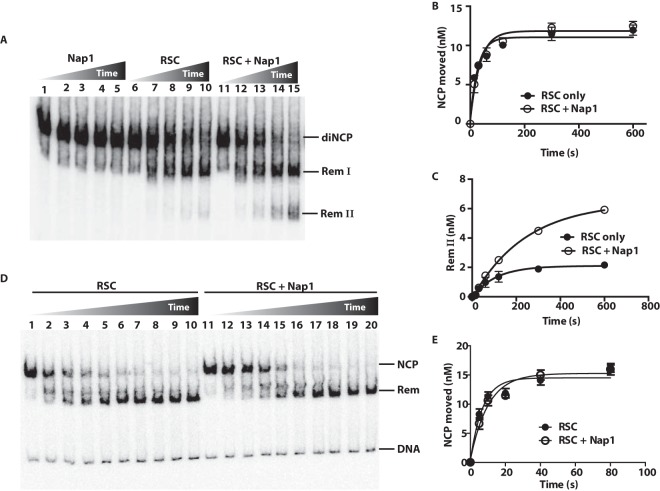
We examined the role of Nap1 in enhancing RSC-mediated disassembly of adjacent nucleosomes in dinucleosomes at moderate concentrations of Nap1. We used a 15-fold (300 nM) molar excess of Nap1 (calculated as a monomer) relative to the amount of RSC or dinucleosome substrate compared to the 1,000-fold or more molar excess used previously (13). Dinucleosomes were analyzed using a native gel shift assay that separates the fully assembled dinucleosomes from remodeling intermediates where either a single H2A-H2B heterodimer is displaced (Rem I) or an entire nucleosome is lost (Rem II), as shown previously (9). Nap1 alone could not disassemble dinucleosomes, whereas RSC had some disassembly activity that was enhanced in the presence of Nap1 (Fig. 1A).
FIG 1.
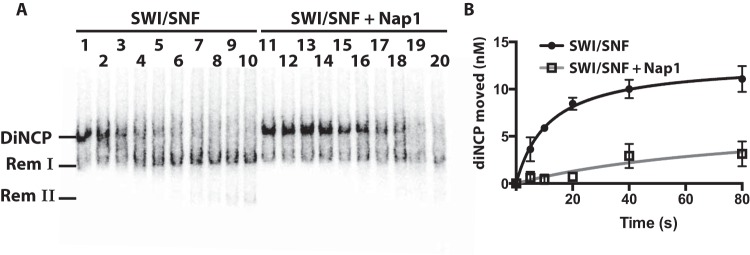
Nap1 promotes nucleosome disassembly of dinucleosomes but not mononucleosomes. (A) The amount dinucleosomes disassembled was tracked by gel shift assays. Reaction mixtures contained 20 nM 601-603 dinucleosomes; 20 nM RSC; and, in lanes 11 to 15, 300 nM Nap1. Reactions were stopped as described in the text at 0 and 40 s and 2, 6, and 20 min. The three bands are labeled, with diNCP being the original dinucleosomes, Rem I being the remodeled dinucleosomes missing one H2A-H2B dimer, and Rem II lacking one of the two nucleosomes, as shown previously (9). (B and C) The extent of dinucleosome movement and the amount of one nucleosome that was displaced or of species II were plotted versus time. The data are the averages from two technical replicates. The error bars represent the standard deviations of the mean. (D) Mononucleosomes were remodeled with RSC in the presence of Nap1 under conditions similar to those for panel A. Mononucleosomes had 29 and 59 bp of DNA flanking the 601 positioning sequence. The times were 0, 5, 10, 20, 40, 80, and 160 s and 5, 10, and 20 min. (E) The amount of mononucleosomes moved by RSC, as evidenced by its disappearance, was plotted versus time in the presence or absence of Nap1 (from panel D).
This disassembly activity was not due to either Nap1 affecting the initial rate of dinucleosome movement mediated by RSC (as determined by the rate of disappearance of the dinucleosome) (Fig. 1B) or the loss of a single H2A-H2B (as seen by the appearance of species I) (data not shown; see Fig. S1 in the supplemental material). However, the presence of Nap1 led to a 3-fold increase in the loss of one of the two nucleosomes (as evidenced by the appearance of species II) (Fig. 1C). Further, Nap1-mediated enhancement of nucleosome disassembly was not observed under comparable conditions using mononucleosomes (Fig. 1D and E). These experiments show that Nap1 can stimulate disassembly of a nucleosome adjacent to a nucleosome being mobilized by RSC and that this stimulation is distinct from the effects of Nap1 and RSC on mononucleosomes.
Nap1 does not enhance the dinucleosome binding or ATPase activities of RSC.
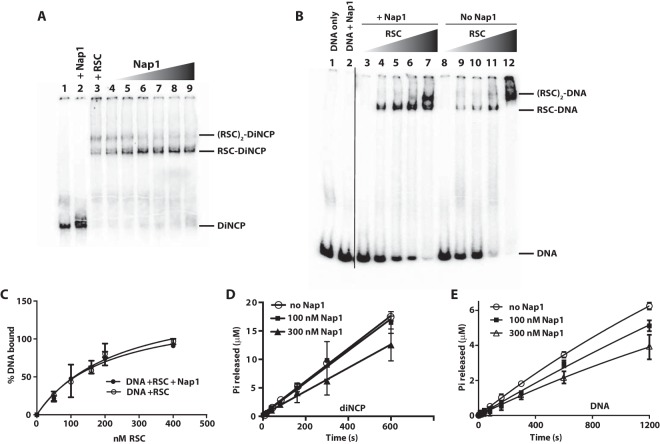
We examined the effects of Nap1 on RSC binding to both dinucleosomes and DNA using native gel shift assays. Notably, under these assay conditions, Nap1 alone did not bind to nucleosomes, DNA, or RSC. In these assays, the complex between RSC and dinucleosomes migrated as two main bands, likely corresponding to dinucleosomal DNA with one and two molecules of RSC bound (Fig. 2A, lane 3). The second molecule of RSC presumably bound nonspecifically to nucleosomes and was readily displaced by Nap1, whereas the other molecule of RSC was specifically bound and retained on nucleosomes even at high Nap1 concentrations of 3 μM (150-fold excess of Nap1 relative to dinucleosome) (Fig. 2A). Similar binding assays were done with a 100-bp DNA, where the addition of Nap1 did not alter the extent of RSC binding (Fig. 2B and C). These data show that Nap1 does not interfere with RSC binding to either nucleosomes or DNA even at 150-fold molar excess.
FIG 2.
Nap1 has only minor effects on RSC binding and ATPase activity with dinucleosomes or DNA. (A and B) The affinity of RSC for dinucleosomes (A) or DNA (B) with and without Nap1 was examined on a native 4% polyacrylamide gel. Samples contained 20 nM RSC, as indicated, as well as 20 nM dinucleosomes (A) or 20 nM 100-bp DNA (B). Concentrations of Nap1 ranging from 30 nM to 2 μM were added (A, lanes 4 to 9) or were held constant at 30 nM (B, lanes 3 to 7). (C) Amount of free DNA bound to RSC with and without Nap1 added plotted versus the concentration of RSC. (D) The rate of ATP hydrolysis was tracked by thin-layer chromatography. Reaction mixtures containing 20 nM dinucleosomes plus 20 nM RSC and no Nap1, 20 nM RSC plus 100 nM Nap1, or 20 nM RSC plus 300 nM Nap1 were plotted versus time. (E) Assays were performed as for panel A, except with 20 nM 100-bp DNA and 40 nM RSC. The data were fitted to a linear regression model using GraphPad Prism. The data are the averages from two technical replicates. The error bars represent the standard deviations of the mean.
To determine whether Nap1 enhances the basic enzymatic properties of RSC, such as ATP hydrolysis and nucleosome movement, we employed well-established ATPase assays using either dinucleosomes or DNA as the activator. We observed that the rate of stimulated ATP hydrolysis of RSC did not increase in proportion to the amount of Nap1 added (Fig. 2D and E). Likewise, there was no significant reduction in ATPase activity in the presence of dinucleosomes and a 5-fold molar excess of Nap1 relative to RSC, but there was a modest yet significant decrease in ATPase activity when Nap1 was present in 15-fold molar excess relative to RSC (Fig. 2D). With free DNA, the reduction of ATPase activity with increasing Nap1 was more pronounced (Fig. 2E); however, this decrease is unlikely to account for Nap1's enhancement of RSC-mediated nucleosome disassembly.
Nap1 enhances RSC-mediated disassembly by altering passage through the adjacent nucleosome.
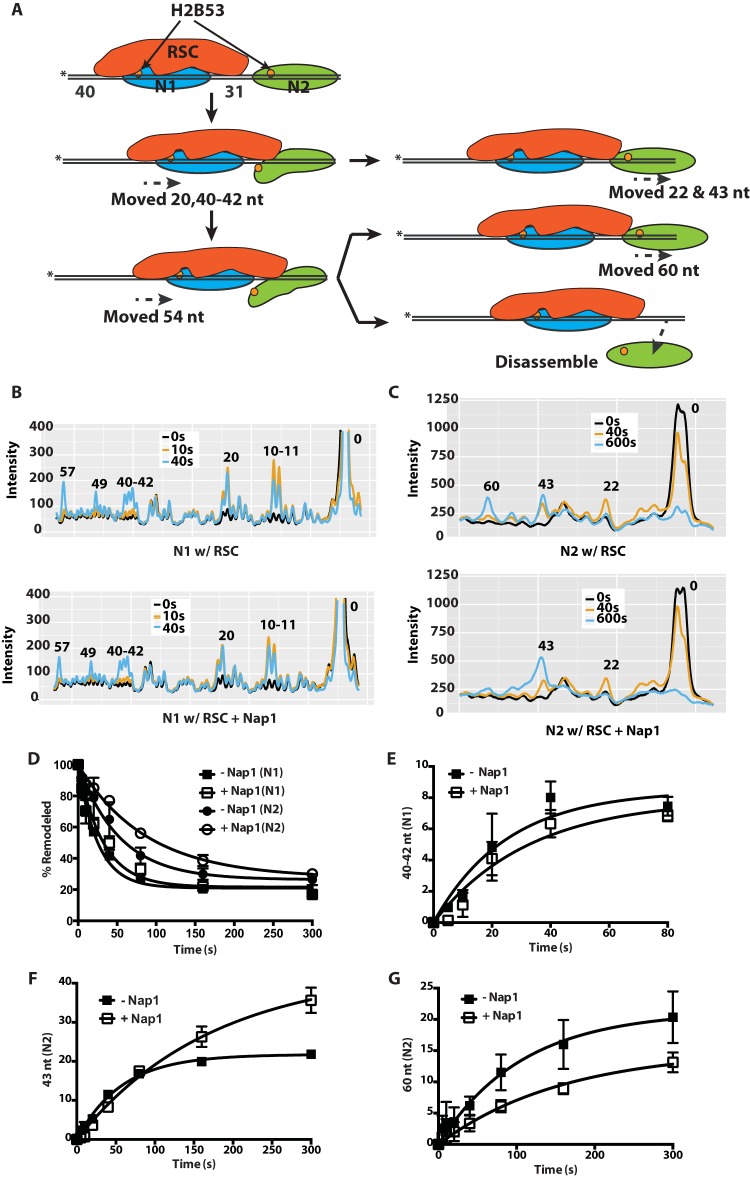
The effect of Nap1 on RSC nucleosome remodeling was examined in more detail by mapping changes in histone-DNA contacts at both nucleosomes in the dinucleosome during remodeling. We tracked the contacts of residue 53 of H2B with nucleosomal DNA by photo-cross-linking, as described previously (9, 30, 31). DNA is cleaved 54 nucleotides (nt) from the dyad axis when residue 53 is cross-linked to DNA (Fig. 3A to C). The nucleosome flanked by 40 and 31 bp of DNA moves first and is referred to as the N1 nucleosome. We found that the N1 nucleosomes moved 10 or 11 nt and then 20 nt from the starting cleavage position toward the second (N2) nucleosome. This initial movement was followed by steps moving the N1 nucleosome in the same direction, 40, 41, or 42; 49; and 57 nt from the starting position (Fig. 3B). Notably, N1 nucleosomes moved away from their initial positions ∼2 times faster than N2 nucleosomes (Fig. 3D).
FIG 3.
Nap1 preferentially affects movement of the second nucleosome in a dinucleosome array. (A) Schematics showing RSC (red) mobilization of the N1 nucleosome (blue) and invasion into an adjacent N2 nucleosome with subsequent eviction (green). The location of the modified residue 53 in histone H2B is shown as an orange circle, and the asterisks indicate the positions of the radiolabel. (B and C) Samples were analyzed on a denaturing 6% polyacrylamide gel as shown in Fig. S2 in the supplemental material, and the gel was scanned with a Fuji FLA-5100. The phosphorimages of three time points, shown as different-color lines, were overlaid. The x and y axes are, respectively, the distance migrated on the gel and the relative band intensity plotted for RSC only and for RSC plus Nap1. The numbers above the peaks indicate the numbers of nucleotides from the dyad axis. The patterns for the N1 (B) and N2 (C) nucleosomes are shown. (D) The extents of movement of N1 and N2 nucleosomes are shown at different times with or without Nap1. (E to G) The amount of N1 nucleosomes moved 40 to 42 nt (E) and the amount of N2 nucleosomes moved 43 and 60 nt (F and G) in the presence or absence of Nap1 were plotted versus time. The data are the averages from two technical replicates. The error bars represent the standard deviations of the mean.
In support of our native gel shift data (Fig. 1), N1 and N2 nucleosome movement proceeded at a similar rates regardless of the presence of Nap1 (Fig. 3B). The rate of movement was the same for each of the 10-, 20-, 31- or 32-, and 40-, 41-, or 42-nt steps for the N1 nucleosomes, confirming that Nap1 does not alter the way in which RSC mobilizes N1 nucleosomes (Fig. 3E; see Fig. S2 in the supplemental material). Initially, as the edge of the N2 nucleosomes closest to N1 moved in response to the movement of N1 nucleosomes, they moved at the same rate regardless of Nap1 (Fig. 3F). However, at the later stages, Nap1 retarded the progression of N2 nucleosomes to the 60-nt position, as evidenced by an ∼2-fold reduction in the rate of movement compared to RSC alone (Fig. 3G). The reduction in N2 nucleosomes that had moved 60 nt was likely because these nucleosomes were disassembled when the DNA from the leading edges of the nucleosomes had been displaced and moved between 43 and 60 nt from their initial positions. These data suggest that Nap1 increases nucleosome loss at the stage where RSC has invaded the adjacent nucleosome close to the interface of H3-H4 and DNA (Fig. 3A). Movement of RSC into the second nucleosome likely exposes H3-H4 to Nap1, allowing binding between them and thereby preventing H3-H4 from rebinding DNA.
Other histone chaperones cannot enhance RSC-mediated disassembly of nucleosomes.
Next, we examined whether Nap1's ability to enhance nucleosome disassembly by RSC was specific to Nap1 or was shared by other histone chaperones. We compared the effect of Nap1 to those of Vps75 and MCM2, which are members of two distinct histone chaperone families. The yeast histone chaperone Vps75 is a structural paralog of Nap1, has high affinity for both H3-H4 and H2A-H2B, and copurifies with the histone H3 acetyltransferase Rtt109 (21, 28, 32, 33). The histone binding domain of MCM2 we used in our tests is more selective for H3-H4 than for H2A-H2B and can hijack interaction sites used by nucleosomal DNA (34). At 15-fold molar excess, neither MCM2 nor Vps75 enhanced nucleosome disassembly. Further, even a 150-fold molar excess of MCM2 had no effect (Fig. 4A and B). Thus, the high-affinity binding between histone chaperones and their histone cargo alone is insufficient to enhance RSC-mediated nucleosome disassembly.
FIG 4.
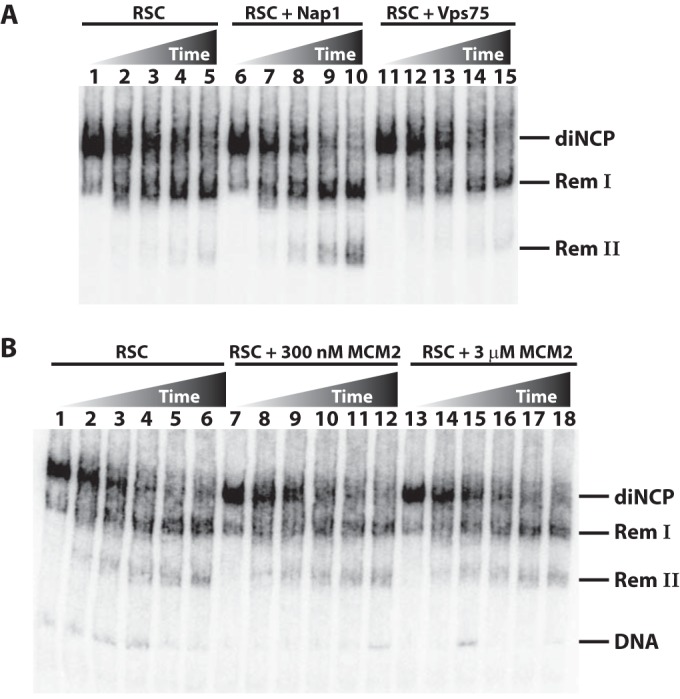
Histone chaperones Vps75 and MCM2 do not affect nucleosome disassembly by RSC. (A) Remodeling assays were performed using 20 nM dinucleosome substrate and 20 nM RSC (lanes 1 to 5), 20 nM RSC plus 300 nM Nap1 (lanes 6 to 10), and 20 nM RSC and 300 nM Vps75 (lanes 11 to 15). Samples were stopped at 0 and 40 s and 2, 6, and 20 min before being analyzed by gel shift. (B) Assays were performed as for panel A, except that they contained 20 nM RSC (lanes 1 to 6), 20 nM RSC plus 300 nM MCM2 (lanes 7 to 12), and 20 nM RSC plus 3 mM MCM2 (lanes 13 to 18). Samples were stopped at 0 and 20 s and 1, 3, 10, and 30 min.
Properties of Nap1 critical for enhancing nucleosome disassembly by RSC.
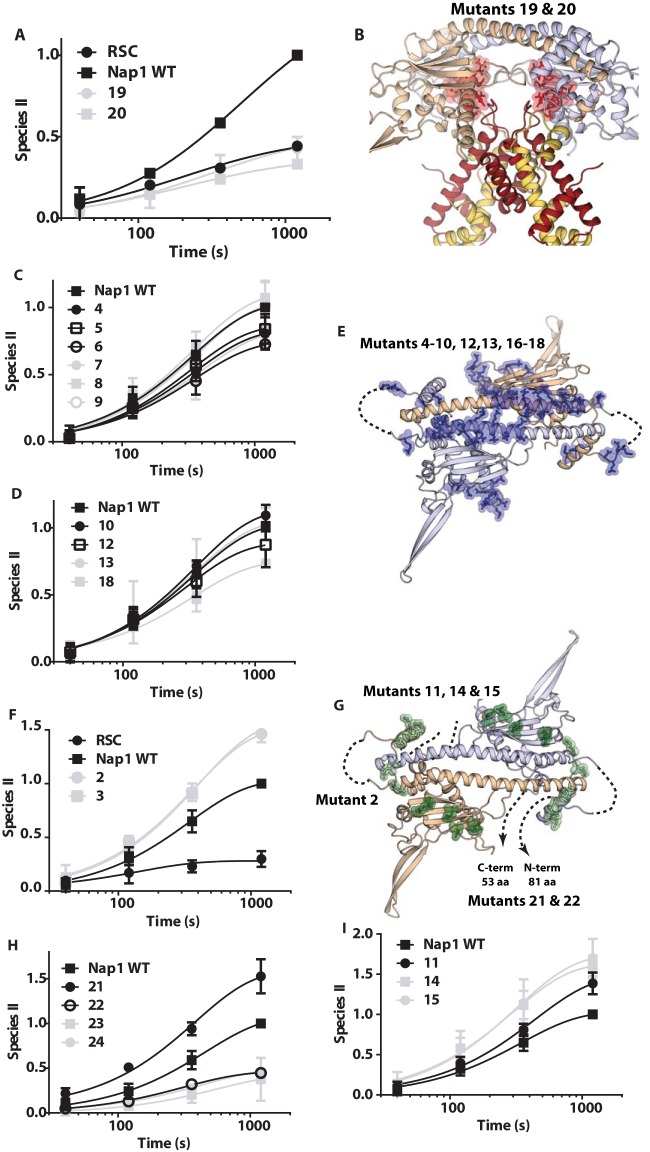
We examined the region(s) of Nap1 important for enhancing RSC-mediated nucleosome disassembly through mutational analysis to better understand the essential properties of Nap1. Previous mutational and structural analyses showed that α-helices 7 and 8 of Nap1 mediate binding to the L2 loop of H2B in an H2A-H2B dimer, with other Nap1 regions potentially binding H2A (25). A total of four deletions and 20 different mutants with multiple amino acid substitutions were used to screen for the region of Nap1 needed for RSC-mediated nucleosome disassembly (see Table S1 in the supplemental material). In the mutants, Nap1 residues were replaced with alanine, glycine, or serine. While most mutations were made to exposed residues to minimize any impact on the Nap1 structure, mutants 20 and 21 involve buried residues. Of the seven point mutations in mutant 20, one residue is buried, while the two point mutations in mutant 21 were purposefully designed to locally destabilize the Nap1 structure. The locations of the mutations and their impacts on Nap1 binding to H2A-H2B are summarized in Table S1 in the supplemental material. Briefly, mutant 2 binds H2A-H2B more tightly than the wild type (see Fig. S4 in the supplemental material), while mutants 19 to 23 bind H2A-H2B less tightly than the wild type, to various degrees (25). Mutants 22 and 23 are N- and C-terminal tail truncations, respectively, while mutants 24 and 25 have compromised oligomerization.
Generally, high-affinity Nap1 binding of H2A-H2B was required for Nap1 enhancement of RSC-mediated nucleosome disassembly (see Table S1 in the supplemental material). Some mutations in Nap1 decreased binding to H2A-H2B by 50- to 60-fold and consequently eliminated enhancement of RSC-mediated disassembly by Nap1 (Fig. 5A, mutants 19 and 20). Mutations that target other regions not critical for H2A-H2B binding, such as in the dimerization helix, the loop and helix of the accessory domain, the 310 helix, and the extended loop region unique to Nap1, did not impact the ability of Nap1 to enhance RSC-mediated disassembly of dinucleosomes (Fig. 5B to D; see Fig. S3B and C in the supplemental material, mutants 4 to 10, 11, 16, and 17). A mutation in the short helix before the dimerization helix increased Nap1 affinity for H2A-H2B and correspondingly increased the ability of Nap1 to enhance RSC-mediated disassembly above that of wild-type Nap1 (Fig. 5F; see Fig. S3A and S4 in the supplemental material, mutant 2). Another mutation in the underside of the dimerization helix also increased Nap1 enhancement of RSC-mediated disassembly over that of wild-type Nap1 (Fig. 5F, mutant 3). Although this mutation did not increase the affinity of Nap1 for H2A-H2B (25), it lies in a region that changes upon H2A-H2B binding. Extensive deletion of the N terminus of Nap1 that reduces Nap1 affinity for H2A-H2B and disrupts dimer formation also inhibits the ability of Nap1 to enhance disassembly (Fig. 5H, mutant 24). These mutations show that Nap1 needs to efficiently bind H2A-H2B and/or H3-H4 in order to enhance nucleosome disassembly by RSC.
FIG 5.
Particular surfaces of Nap1 are critical for RSC-mediated dinucleosome disassembly. (A, C, D, F, H, and I) The rates of RSC-mediated dinucleosome disassembly with WT or mutant Nap1 were examined by gel shift assay as for Fig. 1. Reaction times ranged from 40 s to 20 min. The amount of one nucleosome displaced or the appearance of the remodeled nucleosome II is plotted versus time. The data shown are for Nap1 mutants that do not enhance disassembly (19 and 20) (A), do not change Nap1's ability to enhance disassembly (4 to 10, 12, 13, and 18) (C and D), or enhance more than wild-type Nap1 (2, 3, 11, 14, 15) (F and I) and for deletion mutations of Nap1 (21 to 25) (H). (B, E, and G) Structural models of a Nap1 dimer (wheat and blue) are shown with the positions mutated in Nap1 highlighted (see Table S1 in the supplemental material for details of the mutations). (B) Nap1 is shown bound to two copies of H2A-H2B (yellow and red), with the mutated positions shown as highlighted red sticks corresponding to those in panel A. Only Nap1 is shown in panels E and G, along with the mutated positions corresponding to those in panels C and D with highlighted blue sticks (E) or those in panels F, H, and I with highlighted green sticks (G). Unstructured regions and missing N- and C-terminal tails are indicated by dashed lines.
Given that Nap1 self-associates and forms tetramers at physiological salt concentrations (21, 35–37), we tested whether Nap1 tetramerization was required for Nap1 enhancement of RSC-mediated nucleosome disassembly. Deletion of the β-hairpin (mutant 23) of Nap1 blocks formation of the Nap1 tetramer without interfering with formation of the Nap1 dimer and Nap1 binding to histones (38, 39). We found that deletion of the hairpin of Nap1 limited Nap1's ability to promote RSC-mediated nucleosome disassembly (Fig. 5H).
To test whether the disordered N- and C-terminal tails of Nap1 are also important for RSC-mediated nucleosome disassembly, we deleted the C-terminal tail of Nap1 and found that this abolished the ability of Nap1 to enhance disassembly without drastically reducing the affinity of Nap1 for H2A-H2B (Fig. 5H, mutant 22) (20, 40). Conversely, deletion of the N-terminal tail enhances the effect of Nap1 on RSC-mediated disassembly, also without drastically altering the affinity of Nap1 for H2A-H2B (Fig. 5H, mutant 21). These observations suggest that the N- and C-terminal tails of Nap1 have distinct roles in enhancing RSC-mediated nucleosome disassembly separate from simply binding H2A-H2B. In light of these findings, we reinvestigated the basis of the difference in behavior between Nap1 and Vps75, since the N- and C-terminal tails of Nap1 and Vps75 differ from each other. Nap1 was a much more effective competitor for H2A-H2B binding to free DNA than Vps75 (see Fig. S5 in the supplemental material). Consistent with these differences, when the N- and C-terminal tails of Nap1 were deleted, the ability of Nap1 to compete H2A-H2B from DNA was lost and resembled that of Vps75 (data not shown). Thus, Nap1 could be more selective for enhancing RSC-mediated nucleosome disassembly than Vps75 because it can more effectively compete for H2A-H2B or H3-H4 with DNA.
Two other mutations in the second strand of the β-sheet of Nap1 also increased the efficacy of Nap1 yet did not increase Nap1 affinity for H2A-H2B (Fig. 5G and I, mutants 14 and 15). These mutations may affect interactions between Nap1 and RSC. To test this possibility, we examined the remodeler specificity for Nap1 enhancing nucleosome disassembly. If Nap1 binds uniquely to RSC, then its effects should be limited to RSC and should not be observed with other family members, such as SWI/SNF. Even though Nap1 affinity for H3-H4 and its ability to compete histones from DNA would be advantageous to SWI/SNF-mediated dinucleosome disassembly, if unable to be recruited, then Nap1 would not be able to enhance nucleosome disassembly. We found that Nap1 did not enhance SWI/SNF-mediated nucleosome disassembly even though its mechanism of disassembly is very similar to that of RSC (Fig. 6). In fact, Nap1 had the opposite effect on SWI/SNF-mediated disassembly of dinucleosomes, inhibiting both movement and loss of nucleosomes. The specificity of Nap1 for RSC and particular mutations affecting disassembly without altering its H2A-H2B properties suggests that Nap1 may be recruited by RSC.
FIG 6.
Nap1 interferes with SWI/SNF remodeling of dinucleosomes. (A) Reactions were performed as for Fig. 1, except that 20 nM SWI/SNF was added instead of RSC. The reaction times were 0, 5, 10, 20, 40, 80, and 160 s and 5, 10, and 20 min. (B) The amount of dinucleosomes moved from their starting positions were plotted versus time in the absence and presence of Nap1. The data are the averages from two technical replicates. The error bars represent the standard deviations of the mean.
DISCUSSION
We found that the histone chaperone Nap1 selectively enhanced the nucleosome disassembly activity of RSC on tandem nucleosomes, an activity that is not observed with mononucleosomes. In our dinucleosome system, RSC was preferentially recruited to the nucleosome with the most flanking extranucleosomal DNA, thereby making it possible to track the unidirectional movement of nucleosomes by RSC. In these circumstances, RSC moves one nucleosome into an adjacent nucleosome, causing the edge of the adjacent nucleosome closest to RSC to shift 43 to 60 nt, which most likely promotes disassembly of the nucleosome. Addition of Nap1 did not alter the pattern of DNA movement inside the first nucleosome to be immobilized but did alter the movement of the second nucleosome. The adjacent nucleosome appears to be preferentially lost in the presence of Nap1 only after RSC has invaded 43 to 60 nt into the adjacent nucleosome. This is evident by the absence of adjacent nucleosomes that have moved 60 nt in the presence of Nap1. Because the effect is shown only after invading that far into the adjacent nucleosome, it seems that Nap1 preferentially targets the interface between H3-H4 and DNA when invaded by RSC, thereby promoting nucleosome loss.
Nucleosome loss could potentially be caused by RSC pushing the adjacent nucleosome off the DNA end, with the displaced histones subsequently being captured by the histone chaperone. This possibility seems unlikely given the specificity of Nap1 for enhancing nucleosome disassembly. Other histone chaperones, Vps75 and MCM2, did not affect nucleosome disassembly similarly, even though they bind histones H3-H4 and H2A-H2B. Although the affinity of Nap1 for histones is important, there are clearly other factors that contribute to enhancing nucleosome disassembly, as is evident in the chaperone specificity and from the mutational analysis of Nap1. One critical difference between Vps75 and Nap1 is that Vps75 cannot compete H2A-H2B from DNA. This suggests that the specificity of Nap1 is due in part to its ability to compete H2A-H2B or H3-H4 from DNA rather than RSC merely pushing nucleosomes partially off the DNA. Furthermore, Nap1's affinity for H2A-H2B or H3-H4 and its ability to compete H2A-H2B from DNA are distinct and resolvable from each other, and both are required for nucleosome disassembly. Because disassembly is not increased with mononucleosomes, Nap1 is able to act on DNA-histone interactions only as nucleosomes are destabilized by the intrusion of RSC but will not act on changed DNA-histone interactions that occur as RSC “pumps” DNA through mononucleosomes.
Previous studies using RSC, Nap1, and mononucleosomes have provided contradictory conclusions with respect to the ability of Nap1 to either disassemble mononucleosomes or displace one H2A-H2B dimer to form a hexasome (13, 14). A key difference in these studies was the amount of Nap1 used, 2.4 μM (>1,000-fold excess) for nucleosome disassembly and 43 nM (∼200-fold excess) for hexasome formation. Neither of these results was observed under our conditions. The nucleosome concentration and the type of histones used can both account for these differences. In our study, we used 20 nM nucleosomes, whereas the two previous studies used nucleosome concentrations of 0.2 and 1 nM. We chose a concentration of 20 nM to avoid potential complications due to nucleosome instability upon extensive dilution. We also used yeast histones with yeast Nap1 and RSC to ensure no species variations would influence the outcomes, whereas the prior studies combined yeast RSC with either X. laevis or rat liver histones.
One of the most critical differences between the current study and the earlier studies is that until now, the effects of Nap1 and RSC have not been studied outside the context of a mononucleosome. The colocalization of RSC and Nap1 at promoters and coding regions in yeast, as shown by chromatin immunoprecipitation-sequencing, is consistent with our biochemical evidence that they cooperatively modulate nucleosome dynamics (26, 41–45). Nap1 and RSC associate with actively transcribed genes and promote efficient RNA polymerase II elongation through a nucleosomal template (14, 46). Our data provide evidence that the molecular basis of the enhanced elongation of RNA polymerase II is due to the active displacement of nucleosomes through the concerted actions of RSC and Nap1. RSC is recruited to coding regions through its direct interactions with acetylated histones, either through its multiple bromodomains or by binding directly to RNA polymerase II. RSC recruitment to coding regions is required, in many cases, for active transcription (42). The activities we observed for RSC and Nap1 in our studies indicate how these proteins facilitate early stages of transcription elongation but do not address the question of how these factors might be involved in reassembling chromatin on the template following the passage of the transcription complex (42, 47). It is not known which factors promote Nap1 association with promoters and coding regions, but one possibility suggested by our studies is that RSC helps tether Nap1 to these sites, thereby increasing its efficiency in disassembling nucleosomes that are encountered by the transcription complex. The physical interactions between RSC and Nap1 are likely unstable or transient, as they were not observed to form a stable complex in our gel shift assays. Nonetheless, the interactions between RSC and Nap1 may be important for nucleosome disassembly.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jim Persinger and Briana Dennehey for helpful feedback.
This work was supported by the National Institutes of Health (GM 067777 to K.L. and GM48413 to B.B.). K.L. is also grateful for support from the Howard Hughes Medical Institute.
There are no conflicts of interest
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00195-16.
REFERENCES
- 1.Park YJ, Luger K. 2008. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol 18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das C, Tyler JK, Churchill ME. 2010. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem Sci 35:476–489. doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker PB, Workman JL. 2013. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol 5:a017905. doi: 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. 2004. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev 18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeRoy G, Loyola A, Lane WS, Reinberg D. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J Biol Chem 275:14787–14790. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- 6.Loyola A, Huang JY, LeRoy G, Hu S, Wang YH, Donnelly RJ, Lane WS, Lee SC, Reinberg D. 2003. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol Cell Biol 23:6759–6768. doi: 10.1128/MCB.23.19.6759-6768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson CL. 2009. Reconstitution of nucleosomal arrays using recombinant Drosophila ACF and NAP1. Cold Spring Harb Protoc 2009:pdb.prot5114. doi: 10.1101/pdb.prot5114. [DOI] [PubMed] [Google Scholar]
- 8.Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T. 2003. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol Cell 12:1599–1606. doi: 10.1016/S1097-2765(03)00499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B. 2010. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell 38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee N, Sinha D, Lemma-Dechassa M, Tan S, Shogren-Knaak MA, Bartholomew B. 2011. Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res 39:8378–8391. doi: 10.1093/nar/gkr535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gkikopoulos T, Havas KM, Dewar H, Owen-Hughes T. 2009. SWI/SNF and Asf1p cooperate to displace histones during induction of the Saccharomyces cerevisiae HO promoter. Mol Cell Biol 29:4057–4066. doi: 10.1128/MCB.00400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korber P, Barbaric S, Luckenbach T, Schmid A, Schermer UJ, Blaschke D, Horz W. 2006. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J Biol Chem 281:5539–5545. [DOI] [PubMed] [Google Scholar]
- 13.Lorch Y, Maier-Davis B, Kornberg RD. 2006. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci U S A 103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuryan BG, Kim J, Tran NN, Lombardo SR, Venkatesh S, Workman JL, Carey M. 2012. Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro. Proc Natl Acad Sci U S A 109:1931–1936. doi: 10.1073/pnas.1109994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishimi Y, Yasuda H, Hirosumi J, Hanaoka F, Yamada M. 1983. A protein which facilitates assembly of nucleosome-like structures in vitro in mammalian cells. J Biochem 94:735–744. [DOI] [PubMed] [Google Scholar]
- 16.Lankenau S, Barnickel T, Marhold J, Lyko F, Mechler BM, Lankenau DH. 2003. Knockout targeting of the Drosophila nap1 gene and examination of DNA repair tracts in the recombination products. Genetics 163:611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogner UC, Spyropoulos DD, Le Novere N, Changeux JP, Avner P. 2000. Control of neurulation by the nucleosome assembly protein-1-like 2. Nat Genet 25:431–435. doi: 10.1038/78124. [DOI] [PubMed] [Google Scholar]
- 18.Ohkuni K, Shirahige K, Kikuchi A. 2003. Genome-wide expression analysis of NAP1 in Saccharomyces cerevisiae. Biochem Biophys Res Commun 306:5–9. doi: 10.1016/S0006-291X(03)00907-0. [DOI] [PubMed] [Google Scholar]
- 19.Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. 2005. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J Biol Chem 280:1817–1825. [DOI] [PubMed] [Google Scholar]
- 20.Andrews AJ, Downing G, Brown K, Park YJ, Luger K. 2008. A thermodynamic model for Nap1-histone interactions. J Biol Chem 283:32412–32418. doi: 10.1074/jbc.M805918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowman A, Ward R, Wiechens N, Singh V, El-Mkami H, Norman DG, Owen-Hughes T. 2011. The histone chaperones nap1 and vps75 bind histones h3 and h4 in a tetrameric conformation. Mol Cell 41:398–408. doi: 10.1016/j.molcel.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazurkiewicz J, Kepert JF, Rippe K. 2006. On the mechanism of nucleosome assembly by histone chaperone NAP1. J Biol Chem 281:16462–16472. doi: 10.1074/jbc.M511619200. [DOI] [PubMed] [Google Scholar]
- 23.McBryant SJ, Park YJ, Abernathy SM, Laybourn PJ, Nyborg JK, Luger K. 2003. Preferential binding of the histone (H3-H4)2 tetramer by NAP1 is mediated by the amino-terminal histone tails. J Biol Chem 278:44574–44583. doi: 10.1074/jbc.M305636200. [DOI] [PubMed] [Google Scholar]
- 24.Park YJ, Luger K. 2006. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci U S A 103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Arcy S, Martin KW, Panchenko T, Chen X, Bergeron S, Stargell LA, Black BE, Luger K. 2013. Chaperone Nap1 shields histone surfaces used in a nucleosome and can put H2A-H2B in an unconventional tetrameric form. Mol Cell 51:662–677. doi: 10.1016/j.molcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. 2007. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J 26:2868–2879. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. 2008. Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol 28:6010–6021. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park YJ, Sudhoff KB, Andrews AJ, Stargell LA, Luger K. 2008. Histone chaperone specificity in Rtt109 activation. Nat Struct Mol Biol 15:957–964. doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luger K, Rechsteiner TJ, Richmond TJ. 1999. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol 119:1–16. [DOI] [PubMed] [Google Scholar]
- 30.Kassabov SR, Bartholomew B. 2004. Site-directed histone-DNA contact mapping for analysis of nucleosome dynamics. Methods Enzymol 375:193–210. [DOI] [PubMed] [Google Scholar]
- 31.Kassabov SR, Zhang B, Persinger J, Bartholomew B. 2003. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol Cell 11:391–403. doi: 10.1016/S1097-2765(03)00039-X. [DOI] [PubMed] [Google Scholar]
- 32.Selth L, Svejstrup JQ. 2007. Vps75, a new yeast member of the NAP histone chaperone family. J Biol Chem 282:12358–12362. doi: 10.1074/jbc.C700012200. [DOI] [PubMed] [Google Scholar]
- 33.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. 2007. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell 25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang H, Stromme CB, Saredi G, Hodl M, Strandsby A, Gonzalez-Aguilera C, Chen S, Groth A, Patel DJ. 2015. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat Struct Mol Biol 22:618–626. doi: 10.1038/nsmb.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBryant SJ, Peersen OB. 2004. Self-association of the yeast nucleosome assembly protein 1. Biochemistry 43:10592–10599. doi: 10.1021/bi035881b. [DOI] [PubMed] [Google Scholar]
- 36.Toth KF, Mazurkiewicz J, Rippe K. 2005. Association states of nucleosome assembly protein 1 and its complexes with histones. J Biol Chem 280:15690–15699. doi: 10.1074/jbc.M413329200. [DOI] [PubMed] [Google Scholar]
- 37.Newman ER, Kneale GG, Ravelli RB, Karuppasamy M, Karimi Nejadasl F, Taylor IA, McGeehan JE. 2012. Large multimeric assemblies of nucleosome assembly protein and histones revealed by small-angle X-ray scattering and electron microscopy. J Biol Chem 287:26657–26665. doi: 10.1074/jbc.M112.340422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park YJ, McBryant SJ, Luger K. 2008. A beta-hairpin comprising the nuclear localization sequence sustains the self-associated states of nucleosome assembly protein 1. J Mol Biol 375:1076–1085. doi: 10.1016/j.jmb.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowman A, Hammond CM, Stirling A, Ward R, Shang W, El-Mkami H, Robinson DA, Svergun DI, Norman DG, Owen-Hughes T. 2014. The histone chaperones Vps75 and Nap1 form ring-like, tetrameric structures in solution. Nucleic Acids Res 42:6038–6051. doi: 10.1093/nar/gku232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hieb AR, D'Arcy S, Kramer MA, White AE, Luger K. 2012. Fluorescence strategies for high-throughput quantification of protein interactions. Nucleic Acids Res 40:e33. doi: 10.1093/nar/gkr1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Del Rosario BC, Pemberton LF. 2008. Nap1 links transcription elongation, chromatin assembly, and messenger RNP complex biogenesis. Mol Cell Biol 28:2113–2124. doi: 10.1128/MCB.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spain MM, Ansari SA, Pathak R, Palumbo MJ, Morse RH, Govind CK. 2014. The RSC complex localizes to coding sequences to regulate Pol II and histone occupancy. Mol Cell 56:653–666. doi: 10.1016/j.molcel.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Damelin M, Simon I, Moy TI, Wilson B, Komili S, Tempst P, Roth FP, Young RA, Cairns BR, Silver PA. 2002. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol Cell 9:563–573. doi: 10.1016/S1097-2765(02)00475-6. [DOI] [PubMed] [Google Scholar]
- 44.Ng HH, Robert F, Young RA, Struhl K. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev 16:806–819. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parnell TJ, Huff JT, Cairns BR. 2008. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J 27:100–110. doi: 10.1038/sj.emboj.7601946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carey M, Li B, Workman JL. 2006. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell 24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. 2010. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell 37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.