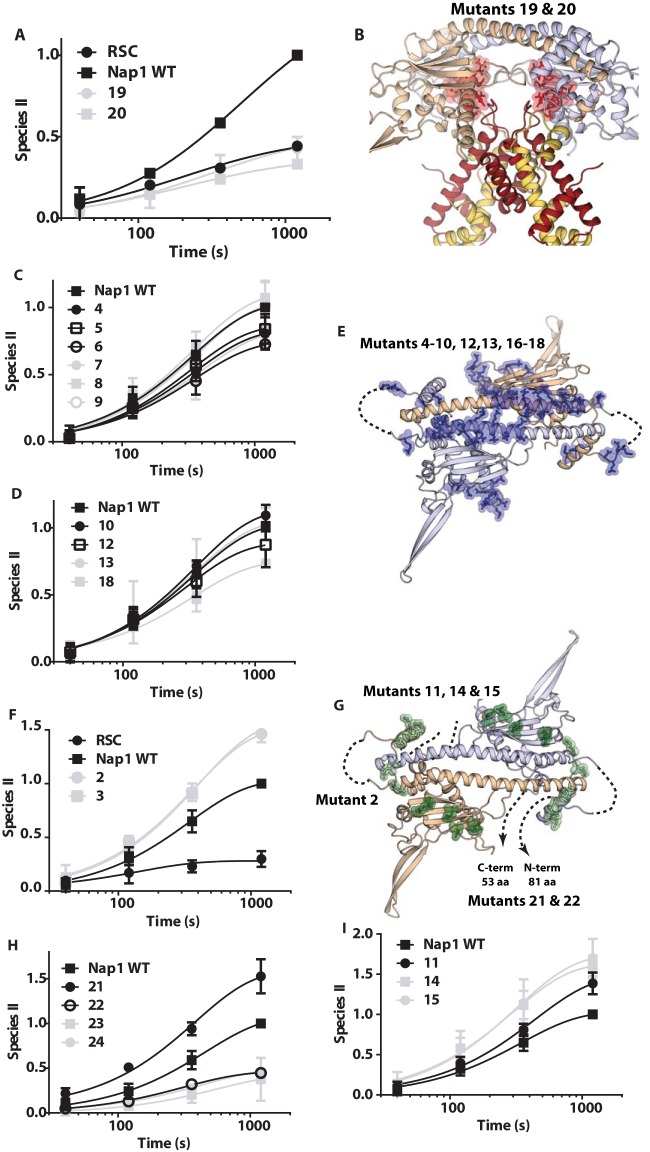
FIG 5.
Particular surfaces of Nap1 are critical for RSC-mediated dinucleosome disassembly. (A, C, D, F, H, and I) The rates of RSC-mediated dinucleosome disassembly with WT or mutant Nap1 were examined by gel shift assay as for Fig. 1. Reaction times ranged from 40 s to 20 min. The amount of one nucleosome displaced or the appearance of the remodeled nucleosome II is plotted versus time. The data shown are for Nap1 mutants that do not enhance disassembly (19 and 20) (A), do not change Nap1's ability to enhance disassembly (4 to 10, 12, 13, and 18) (C and D), or enhance more than wild-type Nap1 (2, 3, 11, 14, 15) (F and I) and for deletion mutations of Nap1 (21 to 25) (H). (B, E, and G) Structural models of a Nap1 dimer (wheat and blue) are shown with the positions mutated in Nap1 highlighted (see Table S1 in the supplemental material for details of the mutations). (B) Nap1 is shown bound to two copies of H2A-H2B (yellow and red), with the mutated positions shown as highlighted red sticks corresponding to those in panel A. Only Nap1 is shown in panels E and G, along with the mutated positions corresponding to those in panels C and D with highlighted blue sticks (E) or those in panels F, H, and I with highlighted green sticks (G). Unstructured regions and missing N- and C-terminal tails are indicated by dashed lines.