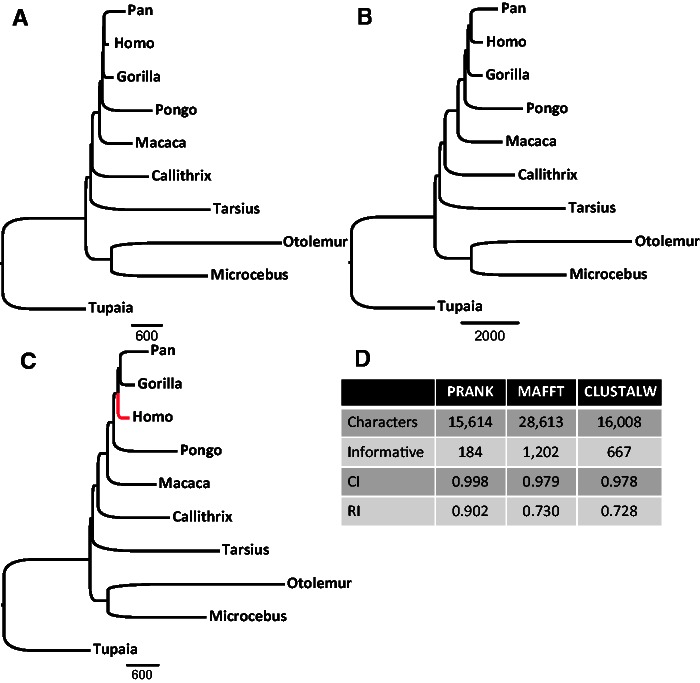
Fig. 5.—
Phylogenetic trees reconstructed using the most reliable indels characters coded from MSAs produced by (A) PRANK, (B) MAFFT, and (C) CLUSTALW and filtered by the RELINDEL method. The correct primate phylogeny was reconstructed when using indels derived from both PRANK and MAFFT. Homo is misplaced in the tree reconstructed based on CLUSTALW MSAs (the erroneous branch is marked in red). Additional statistical information is provided in panel (D) (Informative, number of informative characters; CI, consistence index; RI, retention index).