Abstract
The factors that regulate expression of genes in the 1C family of human cytosolic sulfotransferases (SULT1C) are not well understood. In a recent study evaluating the effects of a panel of transcription factor activators on SULT1C family member expression in LS180 human colorectal adenocarcinoma cells, we found that SULT1C2 expression was significantly increased by 1α,25-dihydroxyvitamin D3 (VitD3) treatment. The objective of our current study was to identify the mechanism responsible for VitD3-mediated activation of SULT1C2 transcription. VitD3 treatment of LS180 cells activated transcription of a transfected luciferase reporter plasmid that contained ∼5 kilobase pairs (kbp) of the SULT1C2 gene, which included 402 nucleotides (nt) of the noncoding exon 1, all of intron 1, and 21 nt of exon 2. Although computational analysis of the VitD3-responsive region of the SULT1C2 gene identified a pregnane X receptor (PXR)-binding site within exon 1, the transfected 5 kbp SULT1C2 reporter was not activated by treatment with rifampicin, a prototypical PXR agonist. However, deletion or mutation of the predicted PXR-binding site abolished VitD3-mediated SULT1C2 transcriptional activation, identifying the site as a functional vitamin D response element (VDRE). We further demonstrated that vitamin D receptor (VDR) can interact directly with the SULT1C2 VDRE sequence using an enzyme-linked immunosorbent assay–based transcription factor binding assay. In conclusion, VitD3-inducible SULT1C2 transcription is mediated through a VDRE in exon 1. These results suggest a role for SULT1C2 in VitD3-regulated physiologic processes in human intestine.
Introduction
The cytosolic sulfotransferases (SULTs) are a family of conjugating enzymes that catalyze the biotransformation of a wide variety of exogenous and endogenous substrates. The human SULT1C subfamily consists of three members: SULT1C2, SULT1C3, and SULT1C4. The characterization of SULT1C substrate specificity, expression, and regulation is incomplete, but available evidence suggests that these enzymes might play metabolic roles during development (Runge-Morris and Kocarek, 2013). Human SULT1C2 is expressed in multiple tissues, including fetal liver, stomach, and the vitamin D target tissues thyroid, intestine, and kidney (Runge-Morris and Kocarek, 2013). SULT1C2 does not metabolize prototypical SULT substrates but has been shown to sulfonate thyroid hormones and some phenols and to bioactivate the procarcinogen N-hydroxy-2-acetylaminofluorene (Sakakibara et al., 1998; Li et al., 2000; Allali-Hassani et al., 2007).
We recently reported that treatment of LS180 human colorectal adenocarcinoma cells with several nuclear receptor activators, including 3-[3-[N-(2-chloro-3-trifluoromethylbenzyl)-(2,2-diphenylethyl)amino]propyloxy]phenylacetic acid (GW3965, liver X receptor agonist), 3-(2,6-dichlorophenyl)-4-(3′-carboxy-2-chlorostilben-4-yl)oxymethyl-5-isopropylisoxazole (GW4064, farnesoid X receptor agonist), rifampicin (pregnane X receptor [PXR] agonist), or 1α,25-dihydroxyvitamin D3 (VitD3, vitamin D receptor agonist), increased SULT1C2 expression (Rondini et al., 2014). The VitD3-mediated induction was especially pronounced and occurred at the mRNA, protein, and promoter activation levels. It therefore seems likely that VitD3-mediated regulation of SULT1C2 could contribute to VitD3-regulated processes in intestine and possibly other VitD3 target tissues, such as the kidney.
VitD3 regulates gene transcription by activating the ligand-dependent nuclear receptor, vitamin D receptor (VDR), which binds as a heterodimer with retinoid X receptor (RXR) to vitamin D response elements (VDREs) in target genes (Haussler et al., 1997). VDREs are classically direct repeats of AGGTCA separated by three nucleotides (i.e., DR3 motifs). We identified the cis-acting element responsible for VitD3-mediated regulation of SULT1C2 transcription.
Materials and Methods
Cell Culture.
LS180 colorectal adenocarcinoma cells were obtained from the American Type Culture Collection (Manassas, VA) and were cultured as previously described elsewhere (Rondini et al., 2014). Human embryonic kidney 293 (HEK293) cells used for protein expression were obtained from Dr. Ye-Shih Ho (Wayne State University, Detroit, MI) and were cultured in Dulbecco’s modified Eagle medium supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum (all purchased from Life Technologies, Grand Island, NY). Cells were maintained under a humidified atmosphere of 95% air, 5% CO2 at 37°C.
Preparation of Firefly Luciferase Reporter Plasmids.
Preparation of the SULT1C2 (−4998:−1)-firefly luciferase (Luc) reporter plasmid has been described previously elsewhere where it was referred to as SULT1C2#2 (Rondini et al., 2014). The SULT1C2 (−4827:−1)-Luc deletion construct was prepared using SULT1C2 (−4998:−1)-Luc as template, HotStar HiFidelity Polymerase (Qiagen, Valencia, CA), and the primer pairs listed in Supplemental Table 1. The polymerase chain reaction fragment was digested with KpnI and NheI and was ligated into the corresponding sites of the pGL4.24 [luc2P/minP] reporter plasmid (Promega, Madison, WI). Site-directed mutagenesis of a computationally predicted (MatInspector; Genomatix, Cincinnati, OH) (Quandt et al., 1995; Cartharius et al., 2005) PXR-binding site was performed using the QuikChange II Kit (Agilent Technologies, Santa Clara, CA) according to manufacturer’s instructions, with SULT1C2 (−4998:−1)-Luc as the template and with the primers listed in Supplemental Table 1.
Transient Transfection Analysis.
Approximately 200,000 LS180 cells were seeded into the wells of 12-well plates. Seventy-two hours later, the cultures were transfected with 1.6 µg of a SULT1C2-Luc reporter plasmid and 1 ng of the Renilla luciferase expression plasmid, pRL-CMV (Promega), as previously described elsewhere (Rondini et al., 2014). After 18 hours, the transfection medium was replaced, and the cells were treated with 0.1% ethanol (Sigma-Aldrich, St. Louis, MO) or 0.1 µM VitD3 (Sigma-Aldrich) for 48 hours. The cells were harvested for measurement of firefly and Renilla luciferase activities using the Dual Luciferase Assay (Promega) and a GloMax Luminometer (Promega).
VDR•VDRE In Vitro Binding Assay.
A VDR expression plasmid was prepared by amplifying the VDR coding sequence (RefSeq NM_000376.2), using LS180 cDNA as template, Herculase II Fusion DNA Polymerase (Agilent Technologies), and the primers listed in Supplemental Table 1. The amplified fragment was digested with HindIII and XhoI and ligated into the pcDNA3.1 expression plasmid (Life Technologies). Approximately 1,000,000 HEK293 cells were seeded into 100-mm plates. On reaching 70%–80% confluency, the cells were transfected with 60 μl Lipofectamine 2000 (Life Technologies), 1.5 µg of VDR-pcDNA3.1, and 1.5 µg RXRα-pSG5 (provided by Dr. Steven Kliewer, University of Texas Southwestern, Dallas, TX). Forty-eight hours later, nuclear proteins were extracted using the NucBuster Protein Extraction kit (Novagen; EMD Millipore, Billerica, MA), and protein concentrations were quantified using the bicinchoninic acid protein assay (Thermo Fisher Scientific, Rockford, IL).
The Universal EZ-TFA Chemiluminescent Transcription Factor Assay (EMD Millipore) was performed according to the manufacturer’s protocol. Briefly, 0.75 µg of HEK293 nuclear extract containing VDR and RXRα, 2 pmol biotinylated capture probe containing the VDRE consensus sequence from the rat osteocalcin gene (also known as bone γ-carboxyglutamate, official symbol Bglap; Markose et al., 1990; Demay et al., 1992), and transcription factor assay buffer were added to the wells of a streptavidin-coated microplate.
Competition for binding of VDR to the capture probe was assessed by adding one of the following unlabeled double-stranded oligonucleotides to the binding reaction: 1) consensus VDRE, 2) mutated consensus VDRE (Gutierrez et al., 2004), 3) SULT1C2 VDRE, or 4) mutated SULT1C2 VDRE. The competitors were added at 1.5- to 50-fold molar excess of the biotinylated capture probe. Background binding was determined by assessing VDR binding to a biotinylated mutated consensus VDRE probe. Capture probe, competitor probe, and negative control probe sequences are listed in Supplemental Table 1. Capture probe-bound VDR was detected using the primary antibody VDR (D-6) X (sc-13133X; Santa Cruz Biotechnology, CA) diluted 1:5000, followed by horseradish peroxidase-conjugated secondary antibody (1:500 dilution). Chemiluminescence was measured using kit reagents and a GloMax Luminometer. Data are expressed relative to VDR•RXR binding to the consensus VDRE capture probe in the absence of a competitor.
Statistical Analysis.
Transfection and transcription factor binding assay data were analyzed using one-way analysis of variance followed by the Newman-Keuls or Dunnett’s multiple comparison test. P < 0.05 was considered statistically significant. Binding curves with 95% confidence intervals were generated using the sigmoidal dose-response algorithm of Prism 6 for Windows (GraphPad Software, La Jolla, CA).
Results and Discussion
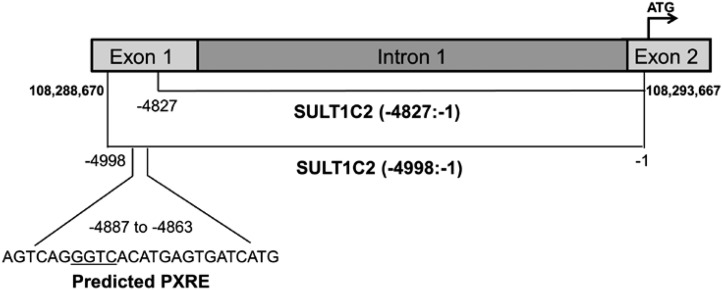
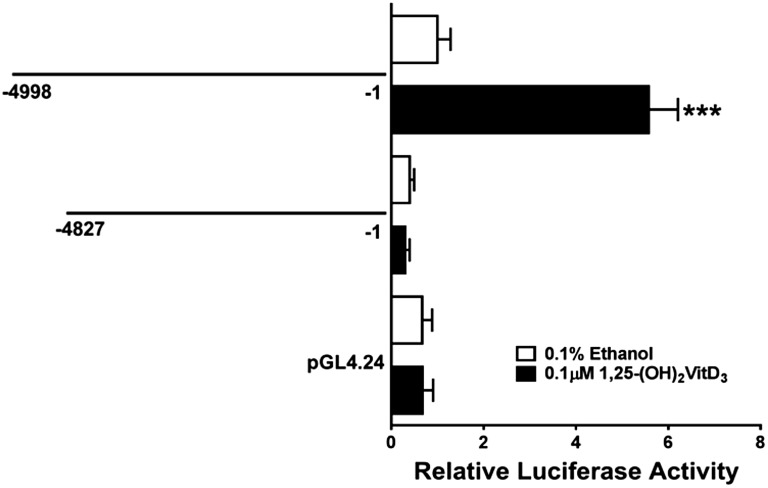
We previously reported that VitD3 treatment of LS180 cells increased activity from a transfected reporter plasmid containing ∼5 kilobase pairs (kbp) of the SULT1C2 gene (−4998:−1 relative to the translation start site in exon 2, shown schematically in Fig. 1) (Rondini et al., 2014). Recently, in a genomewide chromatin immunoprecipitation sequencing analysis of VitD3-treated LS180 cells, Meyer et al. (2012) detected a VDR•RXR binding peak at nucleotides (nt) 108,288,453 to 108,289,105 of chromosome 2 (Gene Expression Omnibus GSE31939; RefSeq NC_000002.12), with the peak center located within the SULT1C2 noncoding exon 1. This information suggested that a VDRE site is located near the 5′-end of our SULT1C2 (−4998:−1) fragment. We therefore deleted 171 nt from the 5′-end of SULT1C2 (−4998:−1), creating the SULT1C2 (−4827:−1)-Luc reporter. Figure 2 shows that this deletion abolished VitD3-mediated SULT1C2 activation, confirming the presence of a VitD3-responsive site in this region.
Fig. 1.
Schematic representation of the SULT1C2 (−4998:−1) fragment. A ∼5 kbp fragment of the SULT1C2 gene containing nt −4998 to −1 relative to the translation start site was amplified and ligated into a luciferase reporter plasmid. This fragment includes 402 nt of the noncoding exon 1, intron 1, and 21 nt of exon 2. Computational analysis identified a putative PXR-binding site (predicted PXR response element, core sequence underlined) at nt −4887 to −4863. SULT1C2 (−4827:−1) shows the PXR-binding site deletion fragment.
Fig. 2.
VitD3 treatment activates reporter expression from SULT1C2 construct (−4998:−1) but not from deletion construct (−4827:−1) in LS180 cells. LS180 cells were transiently transfected with SULT1C2 (−4998:−1)-Luc, SULT1C2 (−4827:−1)-Luc, or empty reporter plasmid (pGL4.24) and treated with 0.1% ethanol or 0.1 µM VitD3 for 48 hours. The cells were harvested, and luciferase activities were measured. Each bar represents the mean ±S.D. of normalized (firefly/Renilla) luciferase activity relative to the activity measured in ethanol-treated SULT1C2 (−4998:−1)-Luc transfected cells (n = 9 wells per group, derived from combining data from three independent experiments with triplicate transfection). ***Significantly different from ethanol-treated cells transfected with the same reporter plasmid, P < 0.001.
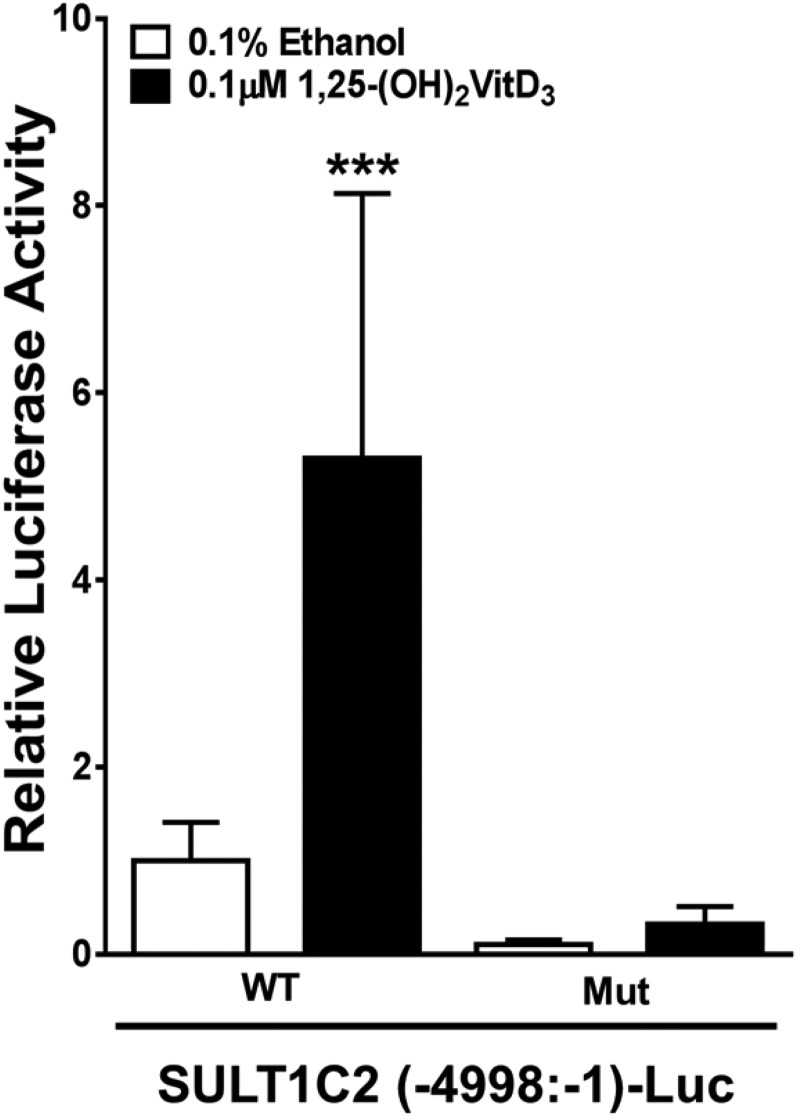
MatInspector software was used to identify putative transcription factor-binding sites within the deleted 171 nt sequence. Instead of detecting a prototypical VDR•RXR DR3 motif in exon 1, a motif identified as a putative PXR•RXR binding site was detected at nt −4887 to −4863 (predicted PXR response element; Fig. 1). However, we previously had reported that treatment of LS180 cells with the prototypical PXR agonist rifampicin did not increase expression from the SULT1C2 (−4998:−1)-Luc reporter (Rondini et al., 2014), suggesting that the computationally-predicted sequence is not a functional PXR response element but rather potentially a VDRE site. Mutation of the core sequence of the predicted PXR-binding site (from GGT to AAC) within the SULT1C2 (−4998:−1)-Luc plasmid caused a 94% reduction in VitD3-mediated SULT1C2 reporter activation compared with the wild-type construct (Fig. 3), further supporting the conclusion that this site is a functional VDRE.
Fig. 3.
Mutation of the predicted PXR-binding site in exon 1 attenuates VitD3-mediated SULT1C2 (−4998:−1)-Luc reporter activation. LS180 cells were transiently transfected with SULT1C2 (−4998:−1)-Luc containing either wild-type (WT) or mutated (Mut) predicted PXR-binding site and treated with 0.1% ethanol or 0.1 µM VitD3. Forty-eight hours later, the cells were harvested and luciferase activities measured. Each bar represents the mean ± S.D. normalized (firefly/Renilla) luciferase activity relative to the activity measured in ethanol-treated WT SULT1C2 (−4998:−1)-Luc transfected cells (n = 6 wells per group, derived from combining data from two independent experiments with triplicate transfection). ***Significantly different from ethanol-treated control, P < 0.001.
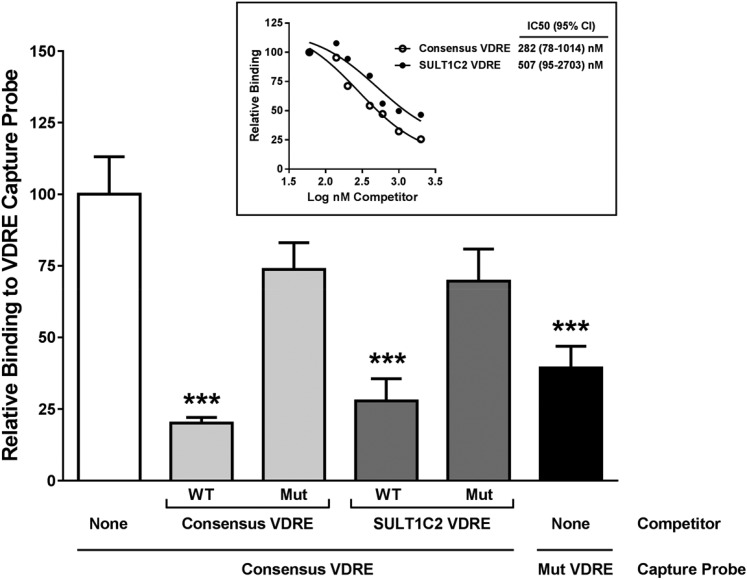
An enzyme-linked immunosorbent assay–based transcription factor-binding assay was used to determine whether VDR•RXR can bind directly to the VDRE site in exon 1 of the human SULT1C2 gene. The capture probe containing the VDRE consensus sequence from the rat osteocalcin gene promoter was incubated with unlabeled competitor probes added in 50-fold molar excess and nuclear protein extract from HEK293 cells expressing VDR and RXRα. The SULT1C2 VDRE competitor significantly inhibited VDR•RXR binding to the capture probe by 74%, which was similar to the inhibition (79%) that was observed when consensus VDRE competitor was added (Fig. 4). Mutated consensus and SULT1C2 VDRE competitors did not significantly inhibit binding to the capture probe (25% and 31% inhibition, respectively). Incubation with increasing amounts (1.5- to 50-fold molar excess) of SULT1C2 VDRE competitor inhibited VDR•RXR binding to the capture probe with an IC50 that was comparable to that of the consensus VDRE competitor (Fig. 4, inset).
Fig. 4.
VDR•RXR binds to the predicted PXR-binding site (VDRE) in exon 1 of SULT1C2. An enzyme-linked immunosorbent assay–based in vitro transcription factor-binding assay was performed as described in Materials and Methods. Binding reactions contained nuclear protein extract and a biotinylated consensus or mutated VDRE capture probe, alone or in the presence of a 50-fold molar excess of either wild-type (WT) or mutated (Mut) consensus VDRE or WT or Mut SULT1C2 VDRE competitor probe. Each bar represents the mean ± S.E.M. binding of VDR•RXR to a VDRE capture probe relative to the binding to the consensus VDRE capture probe that was detected in the absence of competitor (n = 4, derived from the means of four independent binding experiments, each performed with duplicate wells). ***Significantly different from consensus VDRE capture probe in absence of competitor, P < 0.001. Inset: Binding affinity was assessed by adding different amounts (1.5- to 50-fold molar excess) of competitor probes to the consensus VDRE capture probe and nuclear protein extract. IC50 values with 95% confidence intervals (CI) are shown. Each data point is the mean from two independent experiments, each performed in duplicate wells.
VDR and PXR are both members of the NR1I nuclear receptor group and therefore have high sequence similarity (Kliewer et al., 1998). Additionally, they have been shown to recognize similar ligands and regulate similar target genes. For example, VDR and PXR are both activated by the secondary bile acid lithocholic acid (Staudinger et al., 2001; Makishima et al., 2002), and both receptors regulate expression of CYP3A4 (Bertilsson et al., 1998; Schmiedlin-Ren et al., 2001; Thummel et al., 2001). In the case of CYP3A4, PXR and VDR activate transcription through the same cis-acting elements, a proximal everted repeat with six intervening nucleotides motif and a distal DR3 motif (Goodwin et al., 1999; Thummel et al., 2001; Thompson et al., 2002). By comparison, although both PXR and VDR activate SULT1C2 expression in LS180 cells (Rondini et al., 2014), in this case the two receptors do not appear to use the same response element. VitD3 treatment activates SULT1C2 transcription through the element at nt −4887 to −4863, but rifampicin treatment did not increase transcription from the (−4998:−1) reporter construct containing that element (Rondini et al., 2014). Therefore, VDR/PXR regulation of CYP3A4 differs somewhat from SULT1C2 regulation by these two receptors. Presumably the restriction by the SULT1C2 motif to VDR is physiologically important for ensuring appropriate SULT1C2 expression in response to hormonal versus xenobiotic signals. Further research is needed to determine the role that this mode of regulation plays in intestinal physiology.
Supplementary Material
Abbreviations
- DR3
direct repeat of AGGTCA with 3 intervening nucleotides
- GW3965
3-[3-[N-(2-chloro-3-trifluoromethylbenzyl)-(2,2-diphenylethyl)amino]propyloxy]phenylacetic acid
- GW4064
3-(2,6-dichlorophenyl)-4-(3′-carboxy-2-chlorostilben-4-yl)oxymethyl-5-isopropylisoxazole
- HEK
human embryonic kidney cells
- kbp
kilobase pairs
- Luc
firefly luciferase
- nt
nucleotides
- PXR
pregnane X receptor
- RXR
retinoid X receptor
- SULT
cytosolic sulfotransferase
- VitD3
1α,25-dihydroxyvitamin D3
- VDR
vitamin D receptor
- VDRE
vitamin D response element
Authorship Contributions
Participated in research design: Barrett, Kocarek, Runge-Morris.
Conducted experiments: Barrett, Fang.
Performed data analysis: Barrett, Kocarek.
Wrote or contributed to the writing of the manuscript: Barrett, Kocarek, Runge-Morris.
Footnotes
This research was supported by the National Institutes of Health National Institute of Environmental Health Sciences [Grants R01 ES022606 (to M.R.-M.) and Center Grant P30 ES020957].
Part of this work was presented as a poster: [438] Barrett KG, Fang H, Kocarek TA, and Runge-Morris MA, Transcriptional regulation of SULT1C2 by vitamin D receptor in human intestinal and kidney cells. Society of Toxicology 54th Annual Meeting and ToxExpo; 2015 March 22–26; San Diego, California. Society of Toxicology, Reston, VA.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thornton J, Edwards AM, et al. (2007) Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol 5:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Bäckman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. (1998) Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci USA 95:12208–12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933–2942. [DOI] [PubMed] [Google Scholar]
- Demay MB, Kiernan MS, DeLuca HF, Kronenberg HM. (1992) Characterization of 1,25-dihydroxyvitamin D3 receptor interactions with target sequences in the rat osteocalcin gene. Mol Endocrinol 6:557–562. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, Liddle C. (1999) The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol 56:1329–1339. [DOI] [PubMed] [Google Scholar]
- Gutierrez S, Liu J, Javed A, Montecino M, Stein GS, Lian JB, Stein JL. (2004) The vitamin D response element in the distal osteocalcin promoter contributes to chromatin organization of the proximal regulatory domain. J Biol Chem 279:43581–43588. [DOI] [PubMed] [Google Scholar]
- Haussler MR, Haussler CA, Jurutka PW, Thompson PD, Hsieh JC, Remus LS, Selznick SH, Whitfield GK. (1997) The vitamin D hormone and its nuclear receptor: molecular actions and disease states. J Endocrinol 154 (Suppl):S57–S73. [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92:73–82. [DOI] [PubMed] [Google Scholar]
- Li X, Clemens DL, Anderson RJ. (2000) Sulfation of iodothyronines by human sulfotransferase 1C1 (SULT1C1)*. Biochem Pharmacol 60:1713–1716. [DOI] [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. (2002) Vitamin D receptor as an intestinal bile acid sensor. Science 296:1313–1316. [DOI] [PubMed] [Google Scholar]
- Markose ER, Stein JL, Stein GS, Lian JB. (1990) Vitamin D-mediated modifications in protein-DNA interactions at two promoter elements of the osteocalcin gene. Proc Natl Acad Sci USA 87:1701–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Goetsch PD, Pike JW. (2012) VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol Endocrinol 26:37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23:4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondini EA, Fang H, Runge-Morris M, Kocarek TA. (2014) Regulation of human cytosolic sulfotransferases 1C2 and 1C3 by nuclear signaling pathways in LS180 colorectal adenocarcinoma cells. Drug Metab Dispos 42:361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge-Morris M, Kocarek TA. (2013) Expression of the sulfotransferase 1C family: implications for xenobiotic toxicity. Drug Metab Rev 45:450–459. [DOI] [PubMed] [Google Scholar]
- Sakakibara Y, Yanagisawa K, Katafuchi J, Ringer DP, Takami Y, Nakayama T, Suiko M, Liu MC. (1998) Molecular cloning, expression, and characterization of novel human SULT1C sulfotransferases that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J Biol Chem 273:33929–33935. [DOI] [PubMed] [Google Scholar]
- Schmiedlin-Ren P, Thummel KE, Fisher JM, Paine MF, Watkins PB. (2001) Induction of CYP3A4 by 1 alpha,25-dihydroxyvitamin D3 is human cell line-specific and is unlikely to involve pregnane X receptor. Drug Metab Dispos 29:1446–1453. [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, et al. (2001) The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA 98:3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PD, Jurutka PW, Whitfield GK, Myskowski SM, Eichhorst KR, Dominguez CE, Haussler CA, Haussler MR. (2002) Liganded VDR induces CYP3A4 in small intestinal and colon cancer cells via DR3 and ER6 vitamin D responsive elements. Biochem Biophys Res Commun 299:730–738. [DOI] [PubMed] [Google Scholar]
- Thummel KE, Brimer C, Yasuda K, Thottassery J, Senn T, Lin Y, Ishizuka H, Kharasch E, Schuetz J, Schuetz E. (2001) Transcriptional control of intestinal cytochrome P-4503A by 1α,25-dihydroxy vitamin D3. Mol Pharmacol 60:1399–1406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.