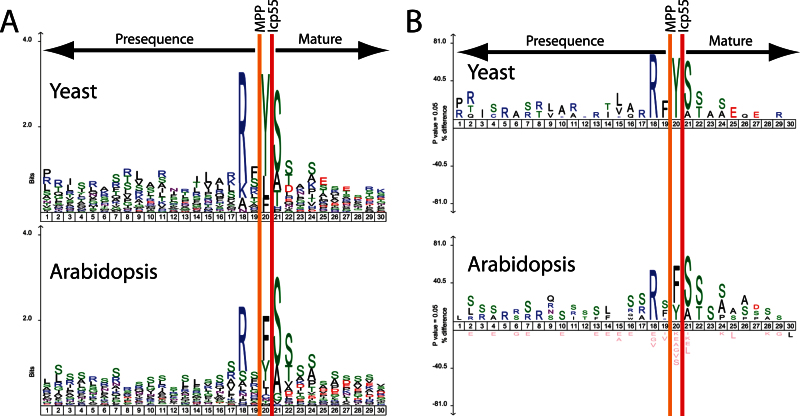
Fig. 2.
SequenceLogo and Icelogo analysis of the cleavage sites of both yeast and Arabidopsis ICP55 proteins. (A) Relative frequency of amino acids in presequences (up to 20 amino acids of the C-terminal segment) and the first 10 amino acids of the mature proteins identified in ICP55 substrates from both yeast and Arabidopsis. (B) The same sequences as in (A), but instead of relative frequency it displays an Icelogo. Icelogo displays the over- and under-represented amino acids compared to each organism’s reference set. The display is in terms of a percentage difference.