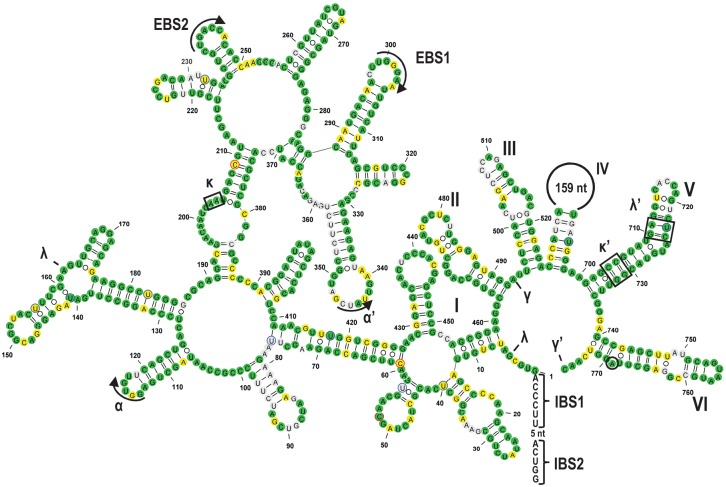
FIGURE 3.
Secondary structure model of rps1i25g2, exemplarily shown for Gleichenia dicarpa, featuring all structural elements typically conserved in group II introns (Michel et al. 1989; Michel and Ferat 1995; Qin and Pyle 1998; Toor et al. 2001; Simon et al. 2008; McNeil et al. 2016). The six intron domains are labeled with Roman numerals (I–VI) and the tertiary interaction sites with Greek letters. Exon binding sites (EBS) and corresponding intron binding sites (IBS) in the 5′ exon are indicated. All nucleotide positions that are identical to the rpl2i846g2 paralog are shown in green. Differences in nucleotide sequences that can be explained by transitions (A/G and C/U) are shown in yellow and all other nucleotides are shown in gray. Experimentally verified RNA editing sites in the intron are marked with blue (C-to-U RNA editing) or red (reverse U-to-C RNA editing) circles. The branch point adenosine for lariat formation is encircled. The intron folding was drawn with the VARNA software (Darty et al. 2009).