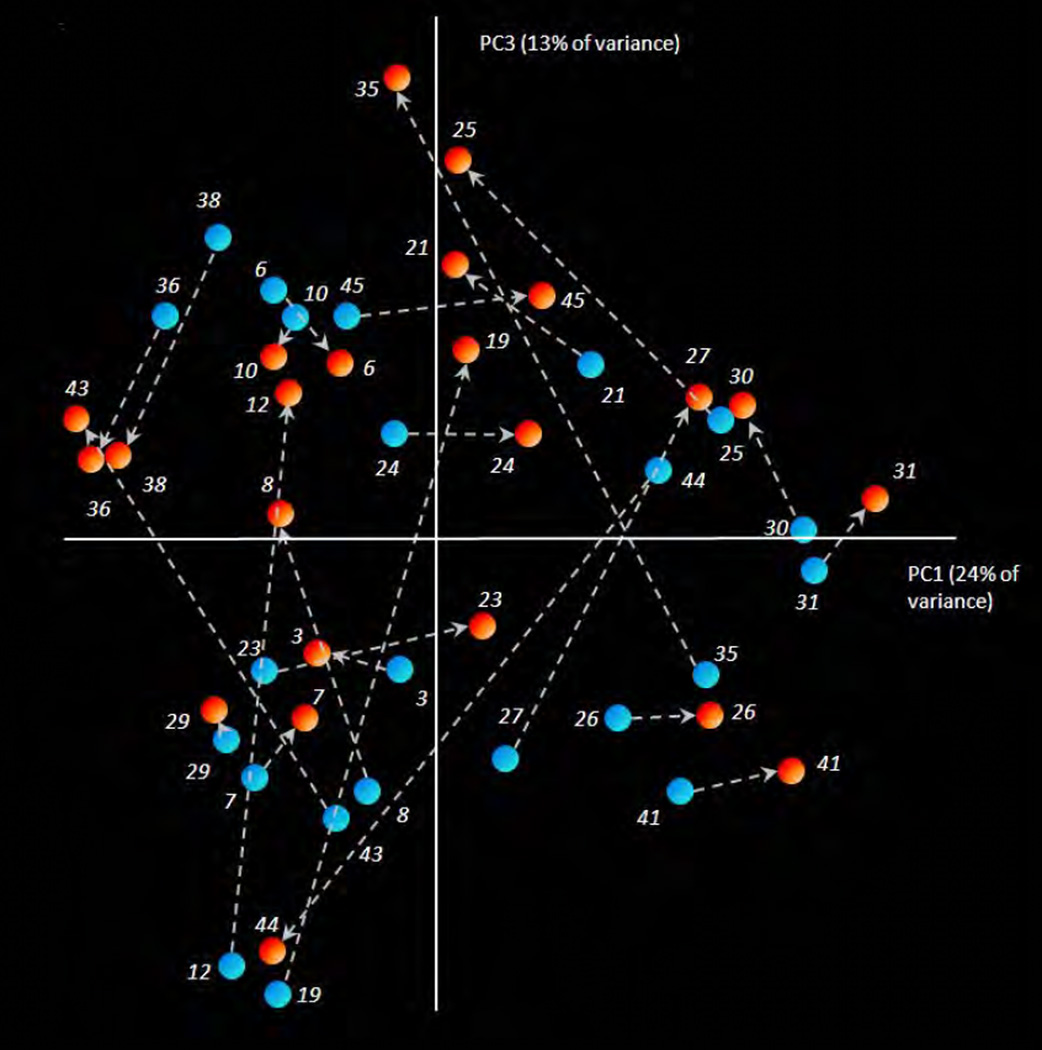
Figure 2.
Principal coordinates analysis (PCoA) plot of the phylogenetic distances (Weighted UniFrac) among the samples from the corresponding pairs of segments (red dots – apical; blue dots – coronal segments) from 23 infected roots. Principal coordinates PC1, PC2 (not shown) and PC3 explained 24%, 17% and 13% of the overall variance among the samples, respectively. The sample pairs (apical and coronal) from the same root are connected with dashed lines. Arrows are pointing towards the apical sample of each pair. The pairs of the samples differ in their phylogenetic relatedness from being very adjacent or similar (roots nr 29 and 10) to very distant or phylogenetically different (roots nr 19, 44, 35 and 12). For the majority of the pairs (16 out of 23) the apical sample was positioned in the positive direction of the PC3 axis, with arrow pointing upwards, indicating the common direction of the microbial shift from coronal to apical microbiome in these samples.