Fig 1. Measuring MHCI protein turnover by 2H2O labeling.
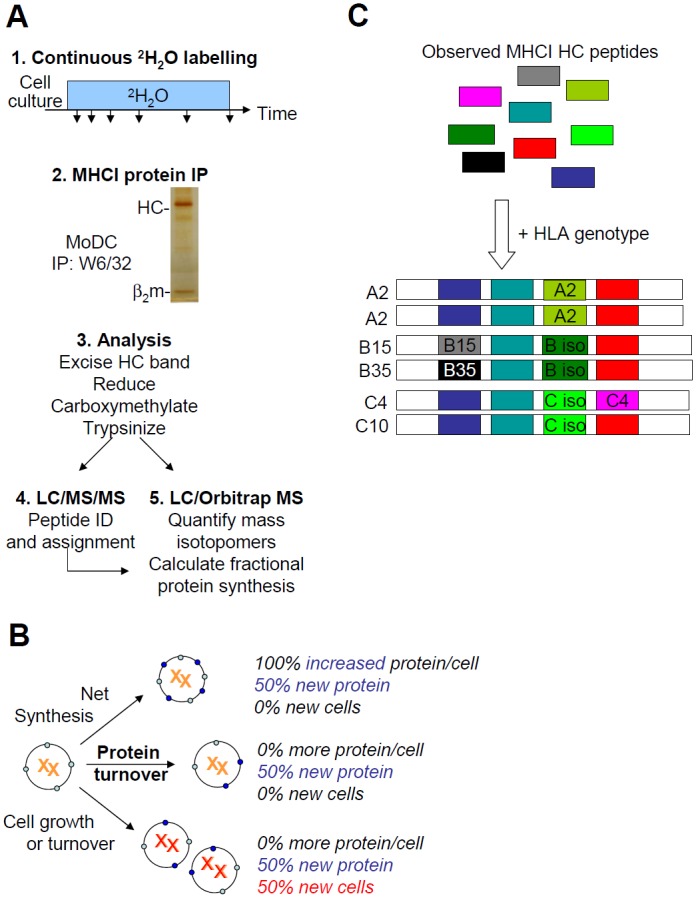
(A) SINEW work flow. See text for details. (B) New protein synthesis may support cell growth, increase net protein levels per cell, or replace protein lost to turnover. These processes contribute additively to protein synthesis. (C) Assignment of peptides to particular MHCI alleles or isotypes. First, LC-MS/MS data (Step 4 in panel A) are screened against sequence databases to identify tryptic fragments derived from any MHCI molecules (top, color-coded boxes). MHCI alleles present in each donor are identified by HLA genotyping, and their predicted amino acid sequences are subjected to tryptic digestion in silico. These virtual digests are compared with each other to identify a subset of peptides that are specific to particular alleles or isotypes (symbolised by boxes with text labels).
