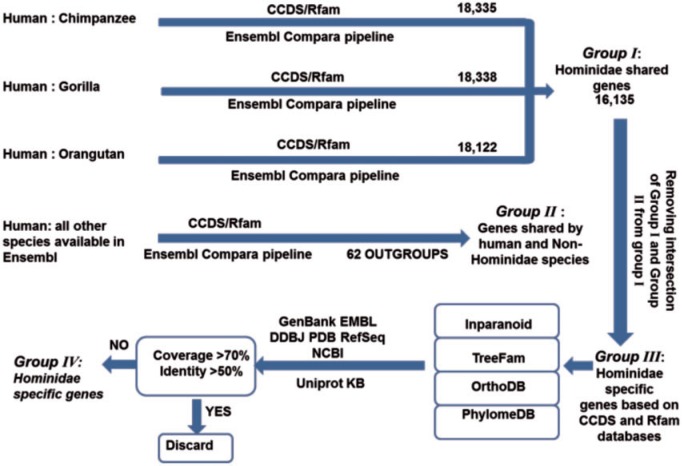
Fig. 1.
Hominidae-specific gene identification pipeline. Human was used as focal species and its genes were searched against the rest of Hominidae members’ genome to identify Hominidae shared genes, indicated by group I. Using the same strategy, pairwise orthologous genes were identified between human and outgroup species, indicated by group II. Intersection of Group I and Group II were omitted from Hominidae shared genes which gives rise to Hominidae-specific genes based on CCDS and Rfam databases. Group III genes were searched in orthology prediction databases (Inparanoid, Treefam, OrthoDB, PhylomeDB) along with DNA and protein databases (Genebank, EMBL, DDBJ, PDB, RefSeq, NCBI and Uniprot KB) and any of the gene queries with significant homology (coverage >70%, identity >50%) in non Hominidae members were discarded.