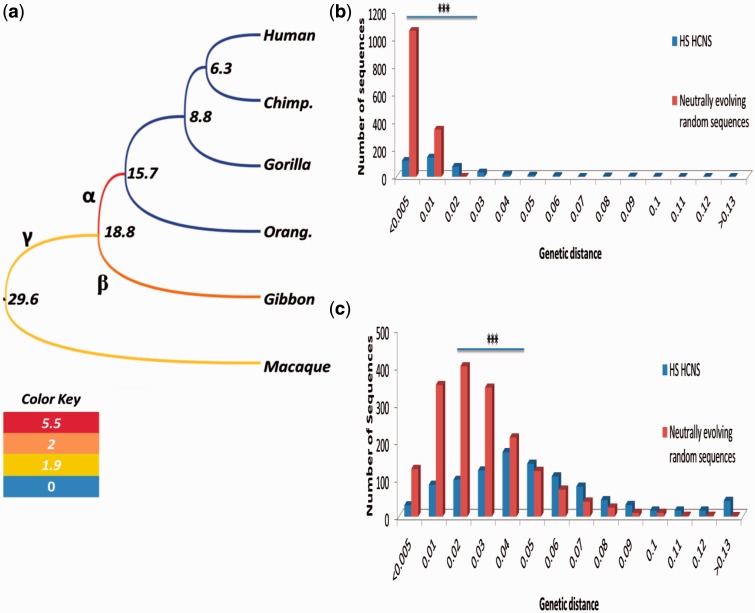
Fig. 4.
HS HCNS substitution rate across catarrhini phylogenetic tree. (a) catarrhini phylogenetic tree color-coded based on the substitution rate per million year in HS HCSNs orthologous sequences. Nucleotide substitution rates in rhesus macaque, gibbon and Hominoidea common ancestor in HS HCNS orthologous sequences are significantly higher than that of neutral evolutionary rate (represented as green in color key). Strongest accelerated mutation rate was observed in Hominidae common ancestor. (b) Comparison of genomic divergence in 32% of HS HCNS’s ancestral sequences in Hominidae common ancestor along with (c) 60% of HS HCNS’s orthologous and ancestral sequences in Hominidae common ancestor and gibbon with that of random sequences under pure neutral evolution reveals signature of accelerated evolution in HS HCNS orthologous and ancestral sequences.