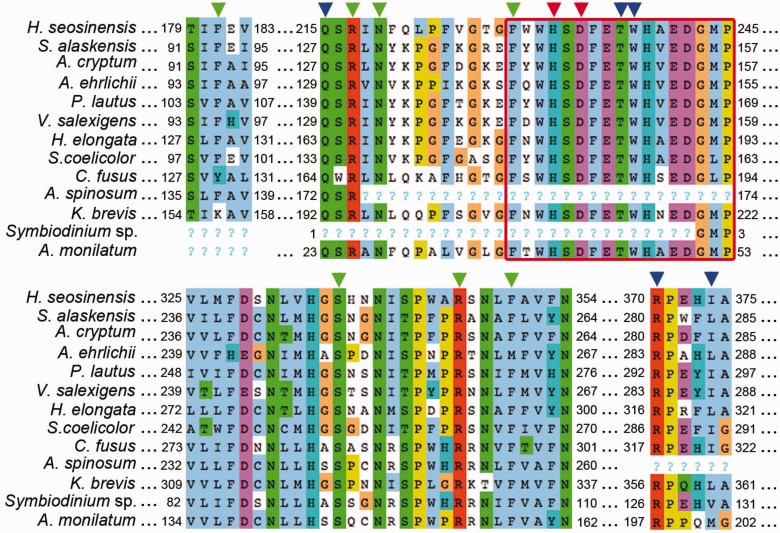
Fig. 6.—
Alignment of ectoine hydroxylase including sequences of characterized enzymes from Sphingopyxis alaskensis (WP_011543221), for which the crystal structure is available, Acidiphilum cryptum (AER00256), Alkalilimnicola ehrlichii (AER00257), Paenibacillus lautus (ACX67869), Virgibacillus salexigens (AAY29689), Halomonas elongata (WP_013333764) and Streptomyces coelicolor (Q93RV9), and sequences from the protists Haloc. seosinensis, Ceratium fusus (CAMPEP_0172939100), Az. spinosum (concatenation of CAMPEP_0180530970, CAMPEP_0180661784 and CAMPEP_0180535134), Karenia brevis (CAMPEP_0178068410), Symbiodinium sp. (CAMPEP_0169646080) and Alexandrium monilatum (CAMPEP_0175754634). Arrowheads indicate residues involved in binding iron (red), 2-oxoglutarate (green), and 5-hydroxyectoine (blue). The consensus sequence of ectoine hydroxylase (FXWHSDFETWHXEDG-M/L-P) is squared in red. “?” indicates missing data for partial sequences.