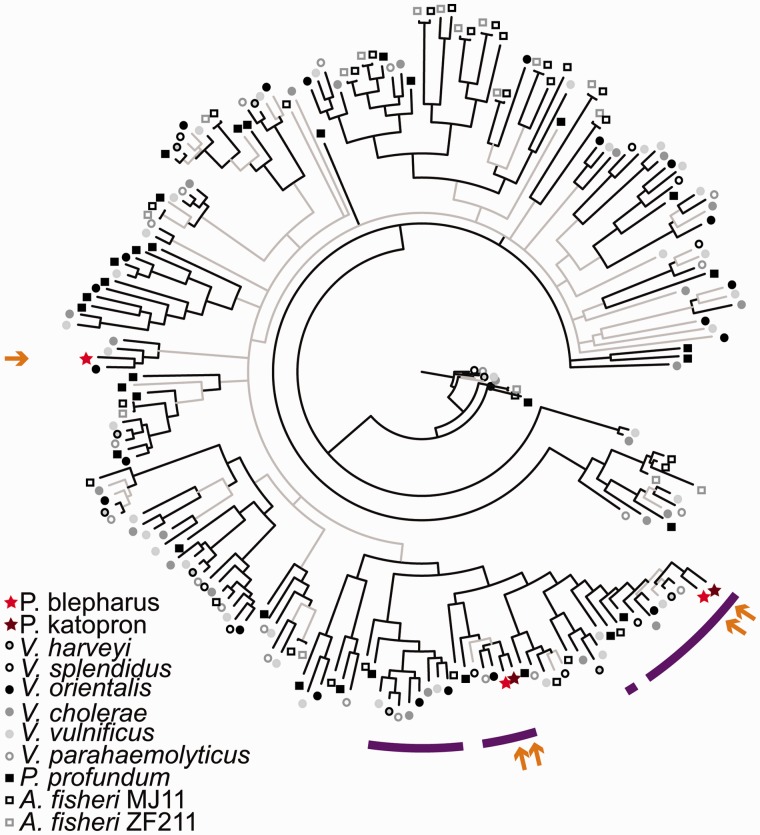
Fig. 3.—
Bayesian tree based on amino acids sequences for all MCP genes found in anomalopid symbionts and the free-living relatives V. harveyi 1DA3, V. splendidus ATCC 33789, V. orientalis CIP102891, V. cholerae O395, V. vulnificus CMCP6, V. parahaemolyticus RIMD 2210633, P. profundum SS9, A. fischeri MJ11, and A. fischeri ZF211. Genes with >40% sequence identity to anomalopid MCP genes, over 40% of the gene length, were included. Branches shown in black have ≥95% posterior probability and branches with less support are in gray. Arrows indicate location of anomalopid symbiont sequences and purple bars denote sequences with a conserved amino acid ligand binding sequence, suggesting that they may respond to amino acids in the environment (Taguchi et al. 1997; Glekas et al. 2010; Nishiyama et al. 2012; Brennan et al. 2013).