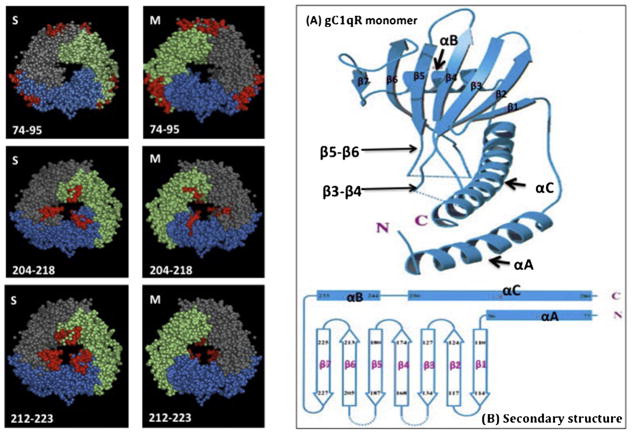
Fig. 10.
Location of the gC1qR residues that disrupt trimer formation.
All the deletions retained the trimeric integrity of the gC1qR molecule with the exception of residues 74–95, 204–218 and 212–223, which did not. The location of these residues (shown in red) within the 3D structure as seen from both the S- and M-faces is shown on the left panel. Figures (A and B) on the right panel show the secondary structure of a single monomer of gC1qR showing that the loops connecting β3 to β4 and β5 to β6 strands occupy a large part of the doughnut hole in the homotrimer (A). In (B) the residues comprising the entire monomer are shown for reference. The structures in A and B were adapted from Jiang et al. (1999).