Figure 2. Neat1 RNA in paraspeckle nuclear bodies: association with paraspeckle proteins and visualization by FISH.
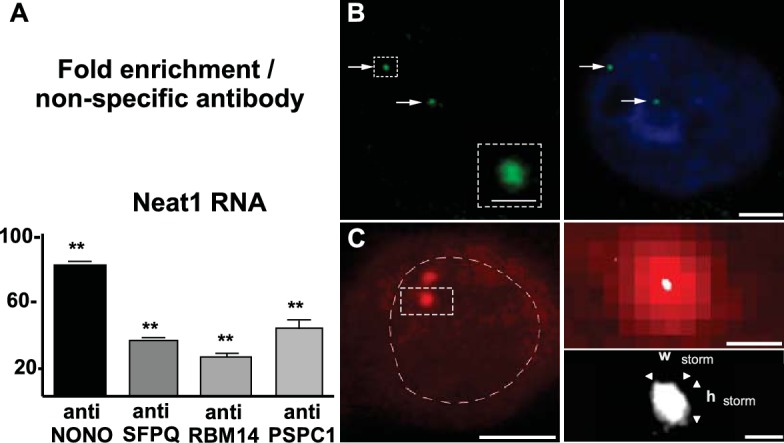
(A) Association of paraspeckle proteins with Neat1 RNA RNA Immuno-Precipitation (RIP) experiments (n=4 for each antibody) using antibodies directed against NONO, SFPQ, RBM14 or PSPC1 show the enrichment in Neat1 RNA obtained as compared to an irrelevant non-specific antibody (see Figure 2—source data 1 for primer sequences). **p<0.01 vs non-specific antibody. B. Visualization of Neat1 RNA by Fluorescence in situ Hybridization and confocal laser scanning microscope Left Panel: RNA-FISH shows the distribution of Neat1 RNA in a few distinct foci (arrows). The round aspect of the foci under confocal laser scanning microscope is shown in the insert in which assigned foci is enlarged. Scale bars: 1 µm Right Panel: The extent of the nucleus is shown with Hoechst staining. Foci containing Neat1 RNA localize within the nucleus sometimes in the close vicinity of nucleus boundaries. Scale bars: 5 µm. (C) Visualization of Neat1 RNA by Fluorescence in situ Hybridization and super resolution Left Panel: Conventional fluorescence microscopy of Neat1 RNA-FISH. The nucleus is outlined with hand-drawn dashed lines to indicate the nuclear periphery. Scale bars: 5 µm. Right Upper Panel: Enlargement of the foci assigned in left panel allows to show the poor resolution of paraspeckle under conventional fluorescence microscopy (in red). In white is the superimposed high-resolution image obtained after STORM analysis. Note that the size of paraspeckle after STORM analysis is strongly reduced. Scale bars: 0.5 µm. Right Bottom Panel: Enlargement of the STORM analysis shown in the upper panel with measurements of width (Wstorm=0.14 µm) and height (hstorm=0.17 µm). Note the elliptical shape of the foci analyzed. Scale bars: 0.1 µm.
DOI: http://dx.doi.org/10.7554/eLife.14837.004
