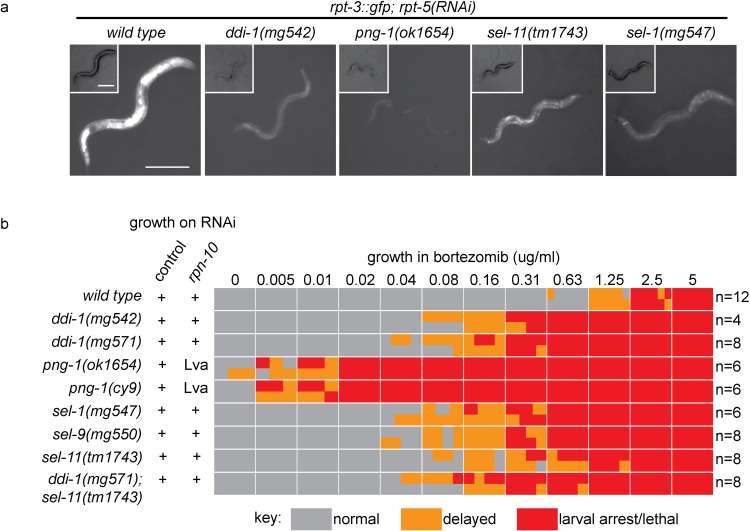
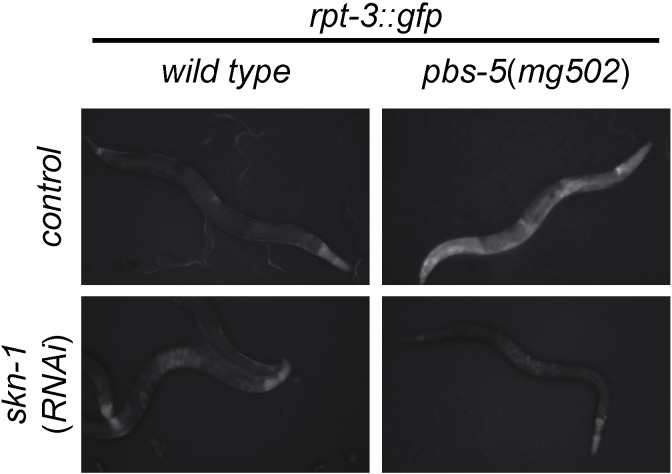
Figure 1. ER-associated degradation factors and the aspartic protease DDI-1 are required for responses to proteasome disruption.
(a) rpt-3::gfp expression following disruption of proteasome function by rpt-5(RNAi) in various mutant backgrounds. Scale bars 100 µm. (b) Table showing growth vs. arrest phenotypes of various mutants in the presence of bortezomib or upon rpn-10 RNAi. For RNAi experiments, L1 animals were incubated for 3 days on indicated RNAi plates, and scored for developmental arrest (+; normal development, Lva; larval arrest). For bortezomib experiments, ~15 L1 animals were incubated for 4 days in liquid cultures containing varying concentrations of bortezomib, and scored for developmental progression. The number (n) of replicate bortezomib experiments performed for each genotype is shown on the right. Each colored rectangle is divided into equal parts to show results from each replicate.